
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,453,694 – 16,453,794 |
| Length | 100 |
| Max. P | 0.704321 |

| Location | 16,453,694 – 16,453,794 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -29.18 |
| Consensus MFE | -21.09 |
| Energy contribution | -22.71 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
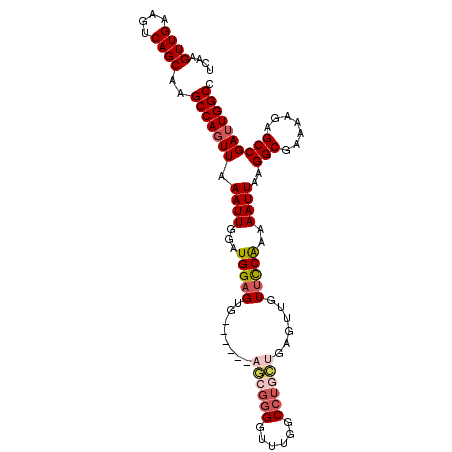
>X_DroMel_CAF1 16453694 100 + 22224390 UCAAGUUGAAGUCAGCAAGCCAGUUAAAUUGGAUGGAGG---UUGGAUGGGGGUUUGGCCUGGUGAGUUGUUCCAA-AAUUAAGGCGAAAAGAGCCGAUUGGCC ....((((....))))..(((((((...(((((...(((---(..(((....)))..))))..........)))))-......(((.......)))))))))). ( -28.52) >DroSec_CAF1 3956 98 + 1 UCAAGUUGAAGUCAGCUAGCCAGUUAAAUUGGCUGGAGUG------CGCGGGGUUUGGCCUGCUGAGUUGUUCCAAAAAUUAAGGCGAAAAGAGCCGAUUGGCC ...(((((....))))).(((((((.((((...((((..(------((((((......)))))...))...))))..))))..(((.......)))))))))). ( -29.30) >DroSim_CAF1 4522 104 + 1 UCAAGUUGAAGUCAGCUAGCCAGUUAAAUUGGCUGGAGUGGAGUGGAGCGGGGUUUGGCCUGCUGAGUUGUUCCAAAAAUUAAGGCGAAAAGAGCCGAUUGGCC ...(((((....))))).(((((((.((((...(((((..(.....((((((......))))))...)..)))))..))))..(((.......)))))))))). ( -34.60) >DroEre_CAF1 4360 91 + 1 UCAAGUUGAAGUCAGCAAGCCAGUUAAAUUGGAUGGAGUG-----------GGGUUGGCUUGGUGAGUUGU-UCG-AAAUUAAGGCGAAAAGAGCCGAGUGGCU ...........(((.(((((((..(..(((......))).-----------.)..))))))).)))((..(-((.-.......(((.......))))))..)). ( -24.30) >consensus UCAAGUUGAAGUCAGCAAGCCAGUUAAAUUGGAUGGAGUG______AGCGGGGUUUGGCCUGCUGAGUUGUUCCAAAAAUUAAGGCGAAAAGAGCCGAUUGGCC ....((((....))))..(((((((.((((...(((((........((((((......))))))......)))))..))))..(((.......)))))))))). (-21.09 = -22.71 + 1.62)


| Location | 16,453,694 – 16,453,794 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -17.47 |
| Consensus MFE | -16.06 |
| Energy contribution | -16.68 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16453694 100 - 22224390 GGCCAAUCGGCUCUUUUCGCCUUAAUU-UUGGAACAACUCACCAGGCCAAACCCCCAUCCAA---CCUCCAUCCAAUUUAACUGGCUUGCUGACUUCAACUUGA ..((((..(((.......)))......-))))......(((.(((((((.............---.................))))))).)))........... ( -15.41) >DroSec_CAF1 3956 98 - 1 GGCCAAUCGGCUCUUUUCGCCUUAAUUUUUGGAACAACUCAGCAGGCCAAACCCCGCG------CACUCCAGCCAAUUUAACUGGCUAGCUGACUUCAACUUGA ..((((..(((.......))).......))))......(((((.(((((........(------(......)).........))))).)))))........... ( -20.03) >DroSim_CAF1 4522 104 - 1 GGCCAAUCGGCUCUUUUCGCCUUAAUUUUUGGAACAACUCAGCAGGCCAAACCCCGCUCCACUCCACUCCAGCCAAUUUAACUGGCUAGCUGACUUCAACUUGA ......((((((.................((((.......(((.((......)).)))....))))....(((((.......)))))))))))........... ( -19.10) >DroEre_CAF1 4360 91 - 1 AGCCACUCGGCUCUUUUCGCCUUAAUUU-CGA-ACAACUCACCAAGCCAACCC-----------CACUCCAUCCAAUUUAACUGGCUUGCUGACUUCAACUUGA ((((....))))................-.((-(....(((.(((((((....-----------..................))))))).))).)))....... ( -15.35) >consensus GGCCAAUCGGCUCUUUUCGCCUUAAUUUUUGGAACAACUCACCAGGCCAAACCCCGCU______CACUCCAGCCAAUUUAACUGGCUAGCUGACUUCAACUUGA ((((....))))..........................(((((((((((.................................)))))))))))........... (-16.06 = -16.68 + 0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:44 2006