
| Sequence ID | X_DroMel_CAF1 |
|---|---|
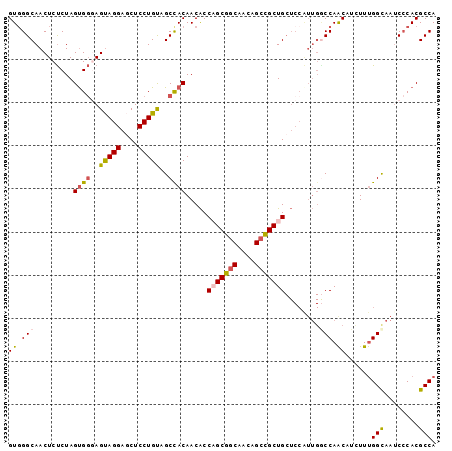
| Location | 16,439,480 – 16,439,606 |
| Length | 126 |
| Max. P | 0.985026 |

| Location | 16,439,480 – 16,439,579 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -37.58 |
| Consensus MFE | -26.78 |
| Energy contribution | -26.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16439480 99 + 22224390 GUGGGCAACUCUCUAGCGGGAGUAGGAGCUCCUGUAGCCACAACACCAGCGGCAACAGCUGCUGCUCCAUUGACCAACAUCUUUGGCAAUCCCACGCCA (((((..((((((....)))))).(((((..((((.(((.(.......).))).)))).....)))))((((.((((.....))))))))))))).... ( -37.30) >DroSec_CAF1 5171 99 + 1 GUGGGCAACUCUCUAGUGGGAGUAGGAGCUCCUGUAGCCACAACACCAGCGGCAACAGCCGCUGCUCCAUUGGCCAACUUUUUUGGCAAUCCCACGCCA (((((..((((((....)))))).(((((...(((....))).....((((((....)))))))))))....(((((.....)))))...))))).... ( -41.40) >DroSim_CAF1 6152 99 + 1 GUGGGCAACUCUCUAGUGGGAGUAGGAGCUCCUGUAGCCACAACACCAGCGCCAACAGCCGCUGCUCCAUUGGCCAACUUUUUUGGCAACCCCACGCCA (((((..((((((....)))))).(((((...(((....))).....((((.(....).)))))))))....(((((.....)))))...))))).... ( -34.50) >DroEre_CAF1 5219 90 + 1 GUGGGCAACUCUCUAGUGGCAGCAGGAACUCCUGCAGCUACAACGCCCGCGGCAACAGCCGCUG---------CCAACAUCUUUGGCAAUCACACGCCA ...(((.........(((((.(((((....))))).))))).......(((((....)))))((---------((((.....)))))).......))). ( -39.00) >DroYak_CAF1 5250 99 + 1 GUUGGCAACUCCCUAGUGCCAGCAGGAACUCCUGCAGCUACAACACCCGCGGCAACAGCCGCUGCUCCGUUGGCCAGCAUCUUCGGUAAUCCCACGCCA ((((((.......((((....(((((....))))).))))((((..(.(((((....))))).)....))))))))))......(((........))). ( -35.70) >consensus GUGGGCAACUCUCUAGUGGGAGUAGGAGCUCCUGUAGCCACAACACCAGCGGCAACAGCCGCUGCUCCAUUGGCCAACAUCUUUGGCAAUCCCACGCCA ((.(((.........((((..(((((....)))))..)))).....(((((((....)))))))........))).))......(((........))). (-26.78 = -26.98 + 0.20)


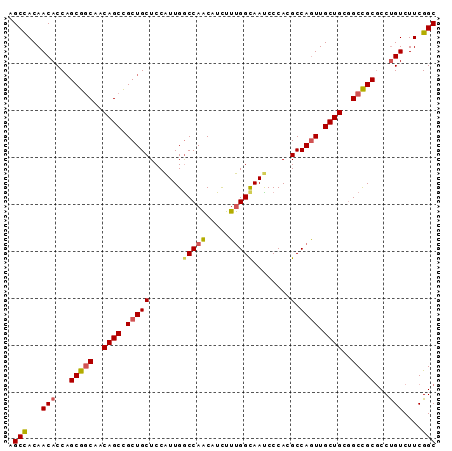
| Location | 16,439,515 – 16,439,606 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -25.45 |
| Energy contribution | -26.01 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16439515 91 + 22224390 AGCCACAACACCAGCGGCAACAGCUGCUGCUCCAUUGACCAACAUCUUUGGCAAUCCCACGCCAGUUGCUGCUGCCGCGCCUGUCUUCGGC .(((...(((...((((((.((((.(((((...((((.((((.....)))))))).....).)))).)))).))))))...)))....))) ( -33.10) >DroSec_CAF1 5206 91 + 1 AGCCACAACACCAGCGGCAACAGCCGCUGCUCCAUUGGCCAACUUUUUUGGCAAUCCCACGCCAGUUGCUGCGGCCGCCCCUGUCUUCGGC .(((...(((...(((((..((((.(((((.......(((((.....)))))........).)))).))))..)))))...)))....))) ( -33.06) >DroSim_CAF1 6187 91 + 1 AGCCACAACACCAGCGCCAACAGCCGCUGCUCCAUUGGCCAACUUUUUUGGCAACCCCACGCCAGUUGCUGCGGCCGCGCCUGUCUUCGGC .(((...(((...((((.....(((((.((...((((((.........(((.....))).)))))).)).))))).)))).)))....))) ( -27.50) >DroYak_CAF1 5285 91 + 1 AGCUACAACACCCGCGGCAACAGCCGCUGCUCCGUUGGCCAGCAUCUUCGGUAAUCCCACGCCAAUUGCUGCGGCUGCGCCCGUCUUCGGC ............((.(((..(((((((.((...((((((......(....).........)))))).)).))))))).)))))........ ( -30.76) >consensus AGCCACAACACCAGCGGCAACAGCCGCUGCUCCAUUGGCCAACAUCUUUGGCAAUCCCACGCCAGUUGCUGCGGCCGCGCCUGUCUUCGGC .(((...(((...(((((..((((.(((((.......(((((.....)))))........).)))).))))..)))))...)))....))) (-25.45 = -26.01 + 0.56)


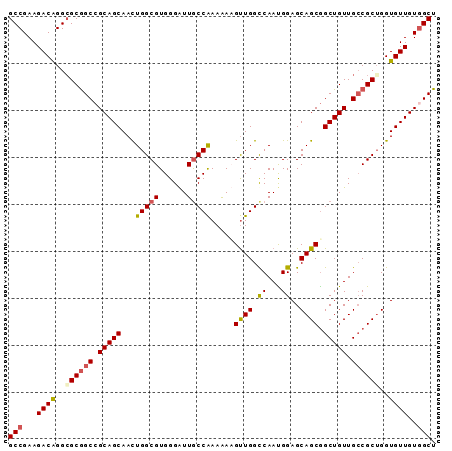
| Location | 16,439,515 – 16,439,606 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 90.29 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -32.80 |
| Energy contribution | -33.42 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16439515 91 - 22224390 GCCGAAGACAGGCGCGGCAGCAGCAACUGGCGUGGGAUUGCCAAAGAUGUUGGUCAAUGGAGCAGCAGCUGUUGCCGCUGGUGUUGUGGCU ((((..((((...(((((((((((...(((((......)))))....(((((.((....)).))))))))))))))))...)))).)))). ( -45.00) >DroSec_CAF1 5206 91 - 1 GCCGAAGACAGGGGCGGCCGCAGCAACUGGCGUGGGAUUGCCAAAAAAGUUGGCCAAUGGAGCAGCGGCUGUUGCCGCUGGUGUUGUGGCU ((((..((((..((((((.(((((...(((((......))))).....((((.((...))..)))).))))).))))))..)))).)))). ( -41.20) >DroSim_CAF1 6187 91 - 1 GCCGAAGACAGGCGCGGCCGCAGCAACUGGCGUGGGGUUGCCAAAAAAGUUGGCCAAUGGAGCAGCGGCUGUUGGCGCUGGUGUUGUGGCU (((.......)))..((((((((((.(..((((......(((((.....)))))((((((........))))))))))..))))))))))) ( -40.10) >DroYak_CAF1 5285 91 - 1 GCCGAAGACGGGCGCAGCCGCAGCAAUUGGCGUGGGAUUACCGAAGAUGCUGGCCAACGGAGCAGCGGCUGUUGCCGCGGGUGUUGUAGCU (((((...(((((((((((((.((..(((((.(((.(((......))).))))))))....)).))))))).)))).))....)))..)). ( -35.60) >consensus GCCGAAGACAGGCGCGGCCGCAGCAACUGGCGUGGGAUUGCCAAAAAAGUUGGCCAAUGGAGCAGCGGCUGUUGCCGCUGGUGUUGUGGCU (((...((((..((((((.(((((...(((((......))))).....((((.((...))..)))).))))).))))))..))))..))). (-32.80 = -33.42 + 0.63)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:37 2006