| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,433,743 – 16,433,849 |
| Length | 106 |
| Max. P | 0.999442 |

| Location | 16,433,743 – 16,433,849 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.53 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -24.09 |
| Energy contribution | -23.57 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
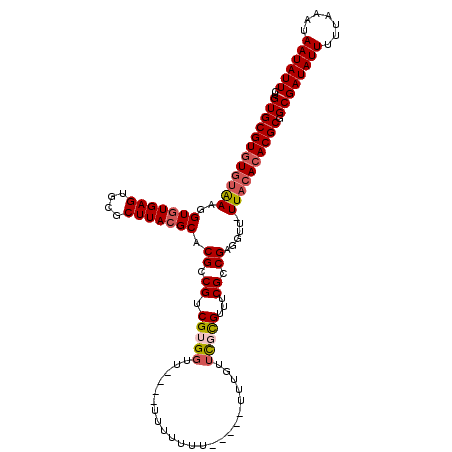
>X_DroMel_CAF1 16433743 106 + 22224390 AAUAUUAUUUAAAAAUAUCGCGGCGUGUGUAA-AACCUCGGCGAAACGCGAACAAA-----A----AA----AACCACGACGGCGUGCGUAAGCGCACUCACACCUUUGCACACGCACAG ......................((((((((((-(.((((((((...)))(......-----.----..----...).))).)).(((((....))))).......))))))))))).... ( -28.50) >DroSec_CAF1 5713 110 + 1 AAUAUUAUUUAAAAAUAUCGCCGCGUGUGUAA-AACCUCGGCGAAACGCGAACAAA-----AAAAAAA----AACCACGACGGCGUGCGUAAGCGCACUCACACCUUUACAUACGCACAG ......................((((((((((-(.((((((((...)))(......-----.......----...).))).)).(((((....))))).......))))))))))).... ( -26.59) >DroSim_CAF1 6722 110 + 1 AAUAUUAUUUAAAAAUAUCGCCGCGUGUGUAA-AACCUCGGCGAAACGCGAACAAA-----AAAAAAA----AACCACGACGGCGUGCGUAAGCGCACUCACACCUUUACACACGCACAG ......................((((((((((-(.((((((((...)))(......-----.......----...).))).)).(((((....))))).......))))))))))).... ( -28.49) >DroEre_CAF1 6057 116 + 1 AAUAUUAUUUAAAAAUAUCGCCGCGUGUGUAAAAAUCUCGACGAUACAAGAAAAAAGAAAUAAAAAAA----AAACACGACGGCGUGCGUAAGCGCACUCACACGUUCAUACACGCACAG ...................(((((((((......(((.....)))..........(....).......----..))))).))))((((((.((((........)))).....)))))).. ( -23.20) >DroYak_CAF1 6199 119 + 1 AAUAUUAUUUAAAAAUAUCGCCGCGUGUGUAA-AAUCUCGGCGGUACGAAAAAAGAGAAAUAAAAAAAAGAAAACCACGACGGCGUGCGUAAGCGCACUCAUACGUUUACACACGCACAG ......................((((((((((-(...(((......))).....(((.................((.....)).((((....)))).))).....))))))))))).... ( -31.20) >consensus AAUAUUAUUUAAAAAUAUCGCCGCGUGUGUAA_AACCUCGGCGAAACGCGAACAAA_____AAAAAAA____AACCACGACGGCGUGCGUAAGCGCACUCACACCUUUACACACGCACAG ......................((((((((((...(((((.....................................))).)).(((((....)))))........)))))))))).... (-24.09 = -23.57 + -0.52)

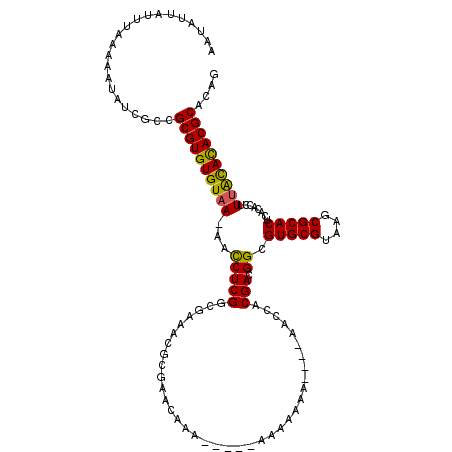

| Location | 16,433,743 – 16,433,849 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.53 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -25.20 |
| Energy contribution | -26.12 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16433743 106 - 22224390 CUGUGCGUGUGCAAAGGUGUGAGUGCGCUUACGCACGCCGUCGUGGUU----UU----U-----UUUGUUCGCGUUUCGCCGAGGUU-UUACACACGCCGCGAUAUUUUUAAAUAAUAUU ..(((((((((.(((((((((((....))))))).((.((.(((((..----..----.-----.....)))))...)).))...))-)).)))))).)))((((((.......)))))) ( -29.70) >DroSec_CAF1 5713 110 - 1 CUGUGCGUAUGUAAAGGUGUGAGUGCGCUUACGCACGCCGUCGUGGUU----UUUUUUU-----UUUGUUCGCGUUUCGCCGAGGUU-UUACACACGCGGCGAUAUUUUUAAAUAAUAUU ..(((((((......(((((....))))))))))))((((.((((((.----......(-----((((...(((...))))))))..-..)).))))))))((((((.......)))))) ( -29.60) >DroSim_CAF1 6722 110 - 1 CUGUGCGUGUGUAAAGGUGUGAGUGCGCUUACGCACGCCGUCGUGGUU----UUUUUUU-----UUUGUUCGCGUUUCGCCGAGGUU-UUACACACGCGGCGAUAUUUUUAAAUAAUAUU ...((((((((((((((((((((....))))))).((.((.(((((..----.......-----.....)))))...)).))...))-)))))))))))..((((((.......)))))) ( -34.04) >DroEre_CAF1 6057 116 - 1 CUGUGCGUGUAUGAACGUGUGAGUGCGCUUACGCACGCCGUCGUGUUU----UUUUUUUAUUUCUUUUUUCUUGUAUCGUCGAGAUUUUUACACACGCGGCGAUAUUUUUAAAUAAUAUU ..(((((((..((.((......)).))..)))))))((((.(((((..----.................(((((......))))).......)))))))))((((((.......)))))) ( -28.97) >DroYak_CAF1 6199 119 - 1 CUGUGCGUGUGUAAACGUAUGAGUGCGCUUACGCACGCCGUCGUGGUUUUCUUUUUUUUAUUUCUCUUUUUUCGUACCGCCGAGAUU-UUACACACGCGGCGAUAUUUUUAAAUAAUAUU ...((((((((((((((.(((.(((((....)))))..)))))).........................(((((......)))))..-)))))))))))..((((((.......)))))) ( -31.10) >consensus CUGUGCGUGUGUAAAGGUGUGAGUGCGCUUACGCACGCCGUCGUGGUU____UUUUUUU_____UUUGUUCGCGUUUCGCCGAGGUU_UUACACACGCGGCGAUAUUUUUAAAUAAUAUU ..((((((((((((..(((((((....))))))).((.((.(((((.......................)))))...)).))......)))))))))).))((((((.......)))))) (-25.20 = -26.12 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:32 2006