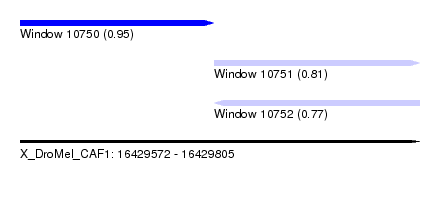
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,429,572 – 16,429,805 |
| Length | 233 |
| Max. P | 0.951519 |

| Location | 16,429,572 – 16,429,685 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.74 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -23.41 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16429572 113 + 22224390 UAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACGGUGGGCAC-------CACAGACUUUUACGGCCCACUGUACGUUCCACCAGUUAGUCAGUCAGUCAUCAAAACAUU ..(((((.((.((...((...((.....((((....))))(((((((((((..-------..............))))))))))).........))....)).)))))))))........ ( -31.19) >DroSec_CAF1 1530 106 + 1 CAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACAGUGGGCAC-------CAAAGACUUUUAGGGCCCACUGUACGUUUCACCAGUCAGUCG-------AUUAAAACAUU ..(((.((((.((...((...((.....((((....))))(((((((((((.(-------(...........)))))))))))))))..))....)))))).-------)))........ ( -34.80) >DroSim_CAF1 2174 106 + 1 CAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACAGUGGGCAC-------CAAAGACUUUUACGGCCCACUGUACGUUUCACCAGUCAGUCG-------AUCAAAACAUU ..(((.((((.((...((...((.....((((....))))(((((((((((..-------.(((....)))...)))))))))))))..))....)))))).-------)))........ ( -34.00) >DroYak_CAF1 2005 109 + 1 UAGAUGGGGUCGAAAAUGUUCACACCAUUCGAUCGUUCGAGUACAGUGGGCACCGGGCACCAAUGGCUUUAAUGGCCCCCUGUUGGUUGCACCAGC----GC-------AUCAAAAUAUU ..((((.(........(((((((...((((((....))))))...)))))))..((((((((((((((.....))))....))))).))).))..)----.)-------)))........ ( -35.60) >consensus CAGAUGGGCUCGAAAAUGUUUACACCAUUCGAUCGUUCGAGUACAGUGGGCAC_______CAAAGACUUUUACGGCCCACUGUACGUUUCACCAGUCAGUCG_______AUCAAAACAUU ....((((.(((((..((....))...))))).)......(((((((((((.......................)))))))))))......))).......................... (-23.41 = -23.98 + 0.56)


| Location | 16,429,685 – 16,429,805 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -24.22 |
| Energy contribution | -24.82 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.813040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16429685 120 + 22224390 GCCAAUUAGUGGGAGUGGGCAAGGGCUGCGCCAGUUCGGAUUUCCAUUGACCCACUGUACGGUGGGUGGGGGAAGUCAAUGCAGAUUGCUUGAAUUUUCUGCUUGGUGGUCUACUUGGGU (((....((((((....(((....))).((((((....(((((((.((.(((((((....))))))).)))))))))...(((((............))))))))))).))))))..))) ( -45.40) >DroSec_CAF1 1636 106 + 1 GCCAAUUAGUGGGAGUGGG-----------CCAGCUCGGAUUUCCAAUGACCCACUGUACGGUGGGUG---GAAGUCAACGCAGAUUGCUUGAAUUUUCUGCUUGGUGGUCUAUUUGGGU .(((.....)))(((((((-----------(((.(...(((((((....(((((((....))))))))---))))))...(((((............)))))..).)))))))))).... ( -40.90) >DroSim_CAF1 2280 106 + 1 GCCAAUUAGUGGGAGUGGG-----------CCAGCUCGGAUUUCCAAUGACCCACUGUACGGUGGGUG---GAAGUCAACGCAGAUUGCUUGAAUUUUCUGCUUGGUGGUCUUUUUGGGU .(((.....)))(((.(((-----------(((.(...(((((((....(((((((....))))))))---))))))...(((((............)))))..).)))))).))).... ( -36.80) >DroEre_CAF1 1860 102 + 1 GCCAAUUUCAAGGC----A-----------AGAGCUCGGAUUUUAAAUAACCCACUGUAAGGUGGGUG---AAACUCUGCGCAGGUUGCUUGAAUUGUCUGCUUGGUGGUCUAUUUUGAU (((((.((((((.(----(-----------(..((.((((.(((.....(((((((....))))))).---))).)))).))...)))))))))........))))).(((......))) ( -27.80) >DroYak_CAF1 2114 103 + 1 GCCAAUUUCAGGGC-UCCG-----------CCAGCUCGAAUUUCAAAUGACCCACUGUACGGUGGGUG---AAAGUCAGCGCAGAUUGCUUCAAUUGUCUGCCUGGUGGUCUAUUUUG-- (((........)))-.(((-----------((((((.((.((((.....(((((((....))))))))---))).)))))((((((..........)))))).)))))).........-- ( -36.50) >consensus GCCAAUUAGUGGGAGUGGG___________CCAGCUCGGAUUUCCAAUGACCCACUGUACGGUGGGUG___GAAGUCAACGCAGAUUGCUUGAAUUUUCUGCUUGGUGGUCUAUUUGGGU ((((...(((((((...................((...((((((.....(((((((....)))))))....))))))...)).((((.....)))))))))))...)))).......... (-24.22 = -24.82 + 0.60)


| Location | 16,429,685 – 16,429,805 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.54 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.28 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.769520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16429685 120 - 22224390 ACCCAAGUAGACCACCAAGCAGAAAAUUCAAGCAAUCUGCAUUGACUUCCCCCACCCACCGUACAGUGGGUCAAUGGAAAUCCGAACUGGCGCAGCCCUUGCCCACUCCCACUAAUUGGC ..((((.(((........(((((............)))))...((.((((...((((((......))))))....)))).)).....(((.((((...)))))))......))).)))). ( -29.10) >DroSec_CAF1 1636 106 - 1 ACCCAAAUAGACCACCAAGCAGAAAAUUCAAGCAAUCUGCGUUGACUUC---CACCCACCGUACAGUGGGUCAUUGGAAAUCCGAGCUGG-----------CCCACUCCCACUAAUUGGC ..((((.(((.....((((((((............))))).))).....---............(((((((((((((....))))..)))-----------))))))....))).)))). ( -29.70) >DroSim_CAF1 2280 106 - 1 ACCCAAAAAGACCACCAAGCAGAAAAUUCAAGCAAUCUGCGUUGACUUC---CACCCACCGUACAGUGGGUCAUUGGAAAUCCGAGCUGG-----------CCCACUCCCACUAAUUGGC ..((((..((.....((((((((............))))).))).....---............(((((((((((((....))))..)))-----------))))))....))..)))). ( -27.30) >DroEre_CAF1 1860 102 - 1 AUCAAAAUAGACCACCAAGCAGACAAUUCAAGCAACCUGCGCAGAGUUU---CACCCACCUUACAGUGGGUUAUUUAAAAUCCGAGCUCU-----------U----GCCUUGAAAUUGGC ..............((((((((..............))))(((((((((---.((((((......))))))............)))))).-----------)----)).......)))). ( -22.66) >DroYak_CAF1 2114 103 - 1 --CAAAAUAGACCACCAGGCAGACAAUUGAAGCAAUCUGCGCUGACUUU---CACCCACCGUACAGUGGGUCAUUUGAAAUUCGAGCUGG-----------CGGA-GCCCUGAAAUUGGC --.........((.(((.(((((............)))))(((((.(((---(((((((......)))))).....)))).)).))))))-----------.)).-(((........))) ( -28.10) >consensus ACCCAAAUAGACCACCAAGCAGAAAAUUCAAGCAAUCUGCGUUGACUUC___CACCCACCGUACAGUGGGUCAUUGGAAAUCCGAGCUGG___________CCCACUCCCACUAAUUGGC ..............(((((((((............)))))(((((.(((....((((((......)))))).....))).))..)))............................)))). (-17.80 = -17.28 + -0.52)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:28 2006