| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,423,898 – 16,424,055 |
| Length | 157 |
| Max. P | 0.796796 |

| Location | 16,423,898 – 16,424,018 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.33 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -23.09 |
| Energy contribution | -23.15 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
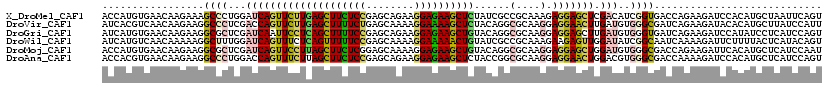
>X_DroMel_CAF1 16423898 120 - 22224390 ACCAUGUGAACAAGAAAGCCCUGGAUCAGUUCUUGAGCUUCUCCGAGCAGAAGGAGAAGCUCUAUCGCCGCAAAGAGGAGCUCGACAUCGGUGACCAGAAGAUCCACAUGCUAAUUCAGU ....((....)).((((((..((((((..((((.((((((((((........))))))))))..((((((....(((...))).....))))))..))))))))))...)))..)))... ( -41.30) >DroVir_CAF1 607 120 - 1 AUCACGUCAACAAGAAGGCCCUCGACCAGUUCUUGAGCUUUUCUGAGCAAAAGGAAAAGCUCUACAGGCGCAAGGAGGAACUUGAUGUGGGCGAUCAGAAGAUACACAUGCUUAUCCAUU ....((((.(((..(((..((((..((.((....((((((((((........)))))))))).)).))(....)))))..)))..))).)))).......((((........)))).... ( -34.40) >DroGri_CAF1 615 120 - 1 AUCAUGUGAACAAGAAGGCGCUCGAUCAAUUCCUCAGCUUUUCCGAGCAGAAGGAGAAGCUGUACAGGCGCAAGGAGGAGCUUGAUGUGGGUGAUCAGAAGAUCCAUAUCCUCAUCCAGU .((.((....)).))..(((((.((.....))..((((((((((........))))))))))....)))))..(((.(((...(((((((((.........)))))))))))).)))... ( -40.90) >DroWil_CAF1 44174 120 - 1 AUCAUGUCAACAAAAAGGCUUUGGAUCAGUUUCUCAGUUUUUCCGAGCAAAAGGAAAAACUGUAUCGCCGCAAAGAAGAGUUGGAUAUCGGCAAUCAAAAGAUUCUUUUACUCAUACAGU ....(((.........(((...(((......)))((((((((((........))))))))))....)))....(((((((((.(((.......)))....)))))))))......))).. ( -26.00) >DroMoj_CAF1 629 120 - 1 ACCAUGUGAACAAGAAGGCGCUCGAUCAGUUCCUUAGCUUCUCGGAGCAAAAGGAGAAGCUGUACAGGCGCAAGGAGGAGCUGGAUGUGGGCGACCAGAAGAUUCACAUGCUCAUCCAAU ..((((((((......((((((((.((((((((((((((((((..........)))))))).......(....)))))))))))...)))))).))......)))))))).......... ( -46.80) >DroAna_CAF1 607 120 - 1 ACCACGUGAACAAGAAGGCCCUGGACCAGUUUCUUAGCUUCUCCGAGCAGAAGGAGAAGCUCUACCGGCGCAAGGAGGAACUGGACGUGGGCGACCAAAAGAUCCACAUGCUCAUCCAGU ....................((((((((((((((((((((((((........))))))))).......(....)))))))))))..((((((........).))))).......))))). ( -43.50) >consensus ACCAUGUGAACAAGAAGGCCCUCGAUCAGUUCCUCAGCUUCUCCGAGCAAAAGGAGAAGCUCUACAGGCGCAAGGAGGAGCUGGAUGUGGGCGACCAGAAGAUCCACAUGCUCAUCCAGU .................((((...((((((((((((((((((((........))))))))))......(....).))))))).)))..))))............................ (-23.09 = -23.15 + 0.06)
| Location | 16,423,938 – 16,424,055 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.11 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -25.22 |
| Energy contribution | -24.17 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16423938 117 - 22224390 ---GAGCUGGAGAAGGCAAACCAGCACCUGAAGAAGUAUAACCAUGUGAACAAGAAAGCCCUGGAUCAGUUCUUGAGCUUCUCCGAGCAGAAGGAGAAGCUCUAUCGCCGCAAAGAGGAG ---(((((((....(((....(((...)))..............((....)).....))).....)))))))..((((((((((........)))))))))).....((.......)).. ( -33.00) >DroVir_CAF1 647 117 - 1 ---GAGCUGGAGAAAGCCAAUCAGCAUCUGAAGAAAUACAAUCACGUCAACAAGAAGGCCCUCGACCAGUUCUUGAGCUUUUCUGAGCAAAAGGAAAAGCUCUACAGGCGCAAGGAGGAA ---(((((((.((..(((..((((...))))...............((.....)).)))..))..)))))))..((((((((((........))))))))))......(....)...... ( -32.60) >DroGri_CAF1 655 117 - 1 ---GAGCUGGAGAAGGCCAAUCAGCAUCUAAAGAAAUACAAUCAUGUGAACAAGAAGGCGCUCGAUCAAUUCCUCAGCUUUUCCGAGCAGAAGGAGAAGCUGUACAGGCGCAAGGAGGAG ---(((((((......)))....((.((....)).......((.((....)).))..)))))).......((((((((((((((........))))))))).......(....)))))). ( -33.60) >DroWil_CAF1 44214 117 - 1 ---GAACUGGAACAGGCCAAUCAGCACUUGAAGAAGUUCAAUCAUGUCAACAAAAAGGCUUUGGAUCAGUUUCUCAGUUUUUCCGAGCAAAAGGAAAAACUGUAUCGCCGCAAAGAAGAG ---(((((((..((((((.....(.((((....)))).).....((....))....))).)))..)))))))..((((((((((........)))))))))).................. ( -27.70) >DroMoj_CAF1 669 117 - 1 ---GAGCUGGAGAAGGCCAAUCAGCAUCUGAAGAAGUACAACCAUGUGAACAAGAAGGCGCUCGAUCAGUUCCUUAGCUUCUCGGAGCAAAAGGAGAAGCUGUACAGGCGCAAGGAGGAG ---..(((((.((..(((..((((...)))).....((((....))))........)))..))..)))))(((((((((((((..........)))))))).......(....)))))). ( -30.70) >DroAna_CAF1 647 119 - 1 UAGGAACUGGAAAAAGCCAAUCAGCU-CUGAAGAAGUACAACCACGUGAACAAGAAGGCCCUGGACCAGUUUCUUAGCUUCUCCGAGCAGAAGGAGAAGCUCUACCGGCGCAAGGAGGAA .(((((((((.....(((..(((...-.)))....((.((......)).)).....)))......))))))))).(((((((((........)))))))))...((..(....)..)).. ( -36.50) >consensus ___GAGCUGGAGAAGGCCAAUCAGCAUCUGAAGAAGUACAACCAUGUGAACAAGAAGGCCCUCGAUCAGUUCCUCAGCUUCUCCGAGCAAAAGGAGAAGCUCUACAGGCGCAAGGAGGAG ...(((((((.....(((..((((...))))..............(....).....)))......)))))))..((((((((((........)))))))))).................. (-25.22 = -24.17 + -1.05)

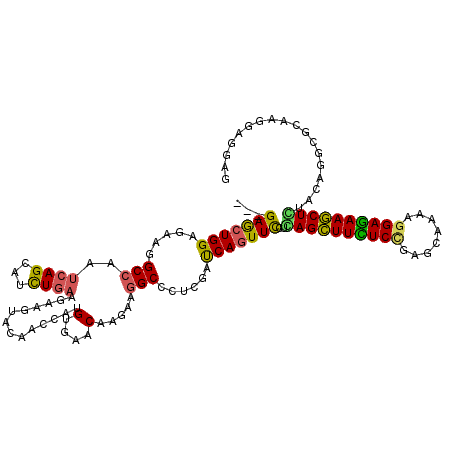
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:24 2006