
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,422,651 – 16,422,851 |
| Length | 200 |
| Max. P | 0.992395 |

| Location | 16,422,651 – 16,422,771 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -46.12 |
| Consensus MFE | -42.70 |
| Energy contribution | -43.08 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16422651 120 - 22224390 CAGUCCCUGGUUGCCCGACCAGCUACUCUCAGCUGGCCACAGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUGGUGCUAAAGCACAUGCCCACGGUGGCCACCUCCAAUUGGC .(((..((((((....))))))..)))...((.(((((((...((((((......))))))(((((.((.(.....).))((((....)))))))))....))))))).))......... ( -41.50) >DroSec_CAF1 43362 120 - 1 CAGUCCCUGGUUGCCCGACCAGCUGCUCUCAGCUGGCCACAGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUGGUGCUGAAGCACAUGCCCACGGUGGCCACCUCCAAUUGGC .(((..((((((....))))))..)))...((.(((((((..(((((((......)))))))((.((((((((.......))))))))))...........))))))).))......... ( -47.40) >DroSim_CAF1 39638 120 - 1 CAGUCCCUGGUUGCCCGACCAGCUGCUCUCAGCUGGCCACAGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUGGUGCUGAAGCACAUGCCCACGGUGGCCACCUCCAAUUGGC .(((..((((((....))))))..)))...((.(((((((..(((((((......)))))))((.((((((((.......))))))))))...........))))))).))......... ( -47.40) >DroYak_CAF1 40793 120 - 1 CCGUCCCUGGUUACCCGACCAGCUGCCCUCAGCUGAUCACGGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUGGUGCUGAAGCACAUGCCCACGGUAGCCACCUCCAACUGGC .......((((((((.((.((((((....)))))).))..(((((((((......)))))).((.((((((((.......))))))))))....)))...))))))))((.......)). ( -48.20) >consensus CAGUCCCUGGUUGCCCGACCAGCUGCUCUCAGCUGGCCACAGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUGGUGCUGAAGCACAUGCCCACGGUGGCCACCUCCAAUUGGC .......((((((((.(.(((((((....))))))).)....(((((((......)))))))((.((((((((.......))))))))))..........))))))))((.......)). (-42.70 = -43.08 + 0.37)

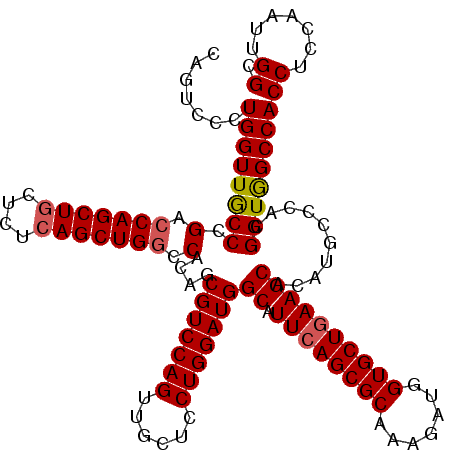
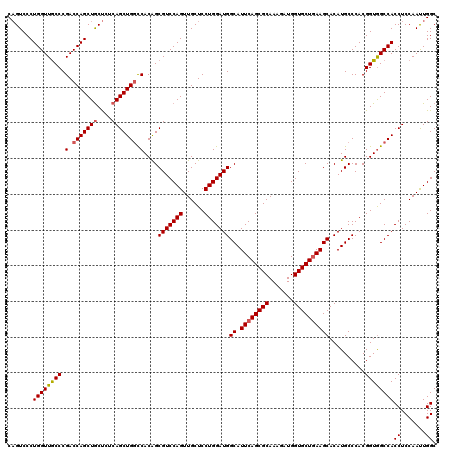
| Location | 16,422,691 – 16,422,811 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.03 |
| Mean single sequence MFE | -45.95 |
| Consensus MFE | -43.95 |
| Energy contribution | -43.45 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16422691 120 + 22224390 CAUCUUUGCGCUGAAUGCCAUCCAGGAGCAACUGGACGCUGUGGCCAGCUGAGAGUAGCUGGUCGGGCAACCAGGGACUGGAGCCUUGAUUGCCAAGCUCUAAGACACCAACAUUUAAUU ..((((.(.(((....((..(((((......))))).))..((((((((((....))))))))))(((((.(((((.(....)))))).))))).))).).))))............... ( -42.80) >DroSec_CAF1 43402 119 + 1 CAUCUUUGCGCUGAAUGCCAUCCAGGAGCAACUGGACGCUGUGGCCAGCUGAGAGCAGCUGGUCGGGCAACCAGGGACUGGAGCCUUGAUUGCCCAGCUCUAAGACACC-ACAUUUAAUU ..((((.(.(((....((..(((((......))))).))...(((((((((....)))))))))((((((.(((((.(....)))))).))))))))).).))))....-.......... ( -48.30) >DroSim_CAF1 39678 119 + 1 CAUCUUUGCGCUGAAUGCCAUCCAGGAGCAACUGGACGCUGUGGCCAGCUGAGAGCAGCUGGUCGGGCAACCAGGGACUGGAGCCUUGAUUGCCCAGCUCUAAGACACC-ACAUUUAAUU ..((((.(.(((....((..(((((......))))).))...(((((((((....)))))))))((((((.(((((.(....)))))).))))))))).).))))....-.......... ( -48.30) >DroYak_CAF1 40833 118 + 1 CAUCUUUGCGCUGAAUGCCAUCCAGGAGCAACUGGACGCCGUGAUCAGCUGAGGGCAGCUGGUCGGGUAACCAGGGACGGGAGCCCUGAUUGCCUAGAUCUAAAGCACU--CAAUUAAUU .........((((((((.(.(((((......))))).).)))..)))))((((.((....((((((((((.(((((.(....)))))).)))))).))))....)).))--))....... ( -44.40) >consensus CAUCUUUGCGCUGAAUGCCAUCCAGGAGCAACUGGACGCUGUGGCCAGCUGAGAGCAGCUGGUCGGGCAACCAGGGACUGGAGCCUUGAUUGCCCAGCUCUAAGACACC_ACAUUUAAUU ..((((.(.(((....((..(((((......))))).))...(((((((((....)))))))))((((((.(((((.(....)))))).))))))))).).))))............... (-43.95 = -43.45 + -0.50)



| Location | 16,422,691 – 16,422,811 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.03 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -36.78 |
| Energy contribution | -37.77 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16422691 120 - 22224390 AAUUAAAUGUUGGUGUCUUAGAGCUUGGCAAUCAAGGCUCCAGUCCCUGGUUGCCCGACCAGCUACUCUCAGCUGGCCACAGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUG ........(((((((((.....(((.((((((((.(((....).)).)))))))).(.((((((......)))))).)..)))((((((......)))))))))).)))))......... ( -41.70) >DroSec_CAF1 43402 119 - 1 AAUUAAAUGU-GGUGUCUUAGAGCUGGGCAAUCAAGGCUCCAGUCCCUGGUUGCCCGACCAGCUGCUCUCAGCUGGCCACAGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUG .......(((-((((((.....((((((((((((.(((....).)).)))))))))(.(((((((....))))))).)..)))((((((......)))))))))))....))))...... ( -46.50) >DroSim_CAF1 39678 119 - 1 AAUUAAAUGU-GGUGUCUUAGAGCUGGGCAAUCAAGGCUCCAGUCCCUGGUUGCCCGACCAGCUGCUCUCAGCUGGCCACAGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUG .......(((-((((((.....((((((((((((.(((....).)).)))))))))(.(((((((....))))))).)..)))((((((......)))))))))))....))))...... ( -46.50) >DroYak_CAF1 40833 118 - 1 AAUUAAUUG--AGUGCUUUAGAUCUAGGCAAUCAGGGCUCCCGUCCCUGGUUACCCGACCAGCUGCCCUCAGCUGAUCACGGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUG ......(((--(((((..........((.(((((((((....).)))))))).)).((.((((((....)))))).))....(((((((......))))))))))))))).......... ( -42.10) >consensus AAUUAAAUGU_GGUGUCUUAGAGCUGGGCAAUCAAGGCUCCAGUCCCUGGUUGCCCGACCAGCUGCUCUCAGCUGGCCACAGCGUCCAGUUGCUCCUGGAUGGCAUUCAGCGCAAAGAUG .......(((.((((((.....((((((((((((.(((....).)).)))))))))(.(((((((....))))))).)..)))((((((......))))))))))))....)))...... (-36.78 = -37.77 + 1.00)


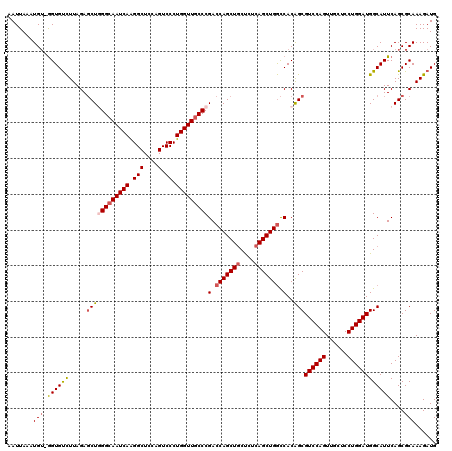
| Location | 16,422,731 – 16,422,851 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -36.64 |
| Consensus MFE | -34.58 |
| Energy contribution | -33.83 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16422731 120 + 22224390 GUGGCCAGCUGAGAGUAGCUGGUCGGGCAACCAGGGACUGGAGCCUUGAUUGCCAAGCUCUAAGACACCAACAUUUAAUUAUAAGAUGCACGUUGUUGAACAAUUUUUUGAGUAAUAAAC .((((((((((....))))))))))(((((.(((((.(....)))))).)))))..((((.((((...(((((....(((....)))......)))))......)))).))))....... ( -34.80) >DroSec_CAF1 43442 119 + 1 GUGGCCAGCUGAGAGCAGCUGGUCGGGCAACCAGGGACUGGAGCCUUGAUUGCCCAGCUCUAAGACACC-ACAUUUAAUUAUAAUAUGCACGUUGUUGAACAAUUUUUUGAGUAAUAAAC (((((((((((....)))))))..((((((.(((((.(....)))))).))))))............))-))....................((((((..(((....)))..)))))).. ( -38.20) >DroSim_CAF1 39718 119 + 1 GUGGCCAGCUGAGAGCAGCUGGUCGGGCAACCAGGGACUGGAGCCUUGAUUGCCCAGCUCUAAGACACC-ACAUUUAAUUAUAAUAUGCACGUUGUUGAACAAUUUUUUGAGUAAUAAAC (((((((((((....)))))))..((((((.(((((.(....)))))).))))))............))-))....................((((((..(((....)))..)))))).. ( -38.20) >DroYak_CAF1 40873 118 + 1 GUGAUCAGCUGAGGGCAGCUGGUCGGGUAACCAGGGACGGGAGCCCUGAUUGCCUAGAUCUAAAGCACU--CAAUUAAUUAUUAAAUGCACGUUGUUGAACAAUAUUUUUCGUAAUAAAC (((((((((((....)))))))))((((((.(((((.(....)))))).)))))).........))...--.......((((((.......((((.....))))........)))))).. ( -35.36) >consensus GUGGCCAGCUGAGAGCAGCUGGUCGGGCAACCAGGGACUGGAGCCUUGAUUGCCCAGCUCUAAGACACC_ACAUUUAAUUAUAAUAUGCACGUUGUUGAACAAUUUUUUGAGUAAUAAAC (((((((((((....)))))))))((((((.(((((.(....)))))).)))))).........))..........................((((((..(((....)))..)))))).. (-34.58 = -33.83 + -0.75)

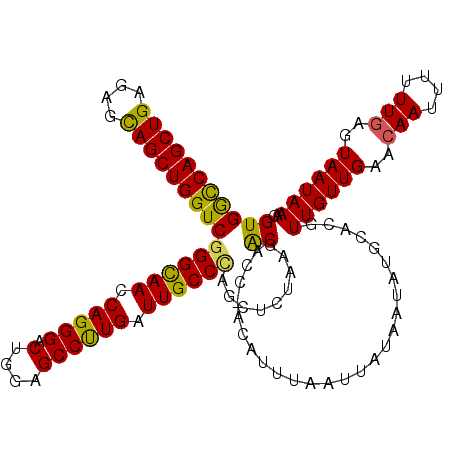
| Location | 16,422,731 – 16,422,851 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -23.16 |
| Energy contribution | -24.73 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16422731 120 - 22224390 GUUUAUUACUCAAAAAAUUGUUCAACAACGUGCAUCUUAUAAUUAAAUGUUGGUGUCUUAGAGCUUGGCAAUCAAGGCUCCAGUCCCUGGUUGCCCGACCAGCUACUCUCAGCUGGCCAC ...................((((.(((....((((.(((....)))))))...)))....))))..((((((((.(((....).)).)))))))).(.((((((......)))))).).. ( -28.10) >DroSec_CAF1 43442 119 - 1 GUUUAUUACUCAAAAAAUUGUUCAACAACGUGCAUAUUAUAAUUAAAUGU-GGUGUCUUAGAGCUGGGCAAUCAAGGCUCCAGUCCCUGGUUGCCCGACCAGCUGCUCUCAGCUGGCCAC ........(((......(((.....)))....((((((.......)))))-)........)))..(((((((((.(((....).)).)))))))))(.(((((((....))))))).).. ( -34.10) >DroSim_CAF1 39718 119 - 1 GUUUAUUACUCAAAAAAUUGUUCAACAACGUGCAUAUUAUAAUUAAAUGU-GGUGUCUUAGAGCUGGGCAAUCAAGGCUCCAGUCCCUGGUUGCCCGACCAGCUGCUCUCAGCUGGCCAC ........(((......(((.....)))....((((((.......)))))-)........)))..(((((((((.(((....).)).)))))))))(.(((((((....))))))).).. ( -34.10) >DroYak_CAF1 40873 118 - 1 GUUUAUUACGAAAAAUAUUGUUCAACAACGUGCAUUUAAUAAUUAAUUG--AGUGCUUUAGAUCUAGGCAAUCAGGGCUCCCGUCCCUGGUUACCCGACCAGCUGCCCUCAGCUGAUCAC (((....((((......)))).....)))..(((((((((.....))))--)))))..........((.(((((((((....).)))))))).)).((.((((((....)))))).)).. ( -32.10) >consensus GUUUAUUACUCAAAAAAUUGUUCAACAACGUGCAUAUUAUAAUUAAAUGU_GGUGUCUUAGAGCUGGGCAAUCAAGGCUCCAGUCCCUGGUUGCCCGACCAGCUGCUCUCAGCUGGCCAC ...................((((........((((...(((......)))..))))....)))).(((((((((.(((....).)).)))))))))(.(((((((....))))))).).. (-23.16 = -24.73 + 1.56)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:22 2006