
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,421,879 – 16,422,030 |
| Length | 151 |
| Max. P | 0.927602 |

| Location | 16,421,879 – 16,421,990 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -20.67 |
| Energy contribution | -20.68 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16421879 111 + 22224390 GCAACAAAUAUUUAAUAAAGUUUAACUAAAGAUCAUUUAUCCGCCGUUACAUCGGCAACAAACUUGGUCGAGGAAU-CAAGUCGCCCGUCCGCAAACUGGACGGAAGAUUUG .........................((...(((((.......((((......))))........))))).))....-((((((..(((((((.....)))))))..)))))) ( -28.26) >DroSec_CAF1 42590 111 + 1 CCAACAAAUAUUUAAUAAAGUUUAACUAAAGAUCAAUUAUCUGCCGUUACAUCGGCAUCAGACUUGGUCGAGGAAC-CAAGUCGCCCGUCCGCAAACUGGACGGAACAUUCG .........................................(((((......)))))...((((((((......))-))))))..(((((((.....)))))))........ ( -30.40) >DroSim_CAF1 38866 111 + 1 CCAACAAAUAUUUAAUAAAGUUUAACUAAAAAUCAAUUGUCUGCCGUUACAUCGGCACCAGACUUGGUCGAGGAAC-CAAGUCGCCCGUCCGCAAACUGGACGGAACAUUCG .........................................(((((......)))))...((((((((......))-))))))..(((((((.....)))))))........ ( -30.30) >DroYak_CAF1 40020 112 + 1 UCAACAAAUAAUUAAUAGAAUUUGACUAAAGACCCUUUAACUGCCGUUACAUUGGCACUGGACUUGGCAGAGGAUUAGAAGUCGGGUGUCCAGAACGUGGACAAAAGAUUCA .................((((((........((((.....((((((...((.......))....))))))..(((.....)))))))(((((.....)))))...)))))). ( -26.00) >consensus CCAACAAAUAUUUAAUAAAGUUUAACUAAAGAUCAAUUAUCUGCCGUUACAUCGGCACCAGACUUGGUCGAGGAAC_CAAGUCGCCCGUCCGCAAACUGGACGGAACAUUCG ..........................................((((......))))....((((((((......)).))))))..(((((((.....)))))))........ (-20.67 = -20.68 + 0.00)


| Location | 16,421,879 – 16,421,990 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -21.17 |
| Energy contribution | -21.05 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.556004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16421879 111 - 22224390 CAAAUCUUCCGUCCAGUUUGCGGACGGGCGACUUG-AUUCCUCGACCAAGUUUGUUGCCGAUGUAACGGCGGAUAAAUGAUCUUUAGUUAAACUUUAUUAAAUAUUUGUUGC (((.((.(((((((.......))))))).)).)))-......((((...(((((((((((......)))).)))))))(((.((((((........)))))).))).)))). ( -26.60) >DroSec_CAF1 42590 111 - 1 CGAAUGUUCCGUCCAGUUUGCGGACGGGCGACUUG-GUUCCUCGACCAAGUCUGAUGCCGAUGUAACGGCAGAUAAUUGAUCUUUAGUUAAACUUUAUUAAAUAUUUGUUGG ((((((((((((((.......))))))..((((((-(((....)))))))))...(((((......)))))..(((((((...)))))))..........)))))))).... ( -34.60) >DroSim_CAF1 38866 111 - 1 CGAAUGUUCCGUCCAGUUUGCGGACGGGCGACUUG-GUUCCUCGACCAAGUCUGGUGCCGAUGUAACGGCAGACAAUUGAUUUUUAGUUAAACUUUAUUAAAUAUUUGUUGG .....((.((((((.......))))))))((((((-(((....)))))))))...(((((......)))))..((((.(((.((((((........)))))).))).)))). ( -36.70) >DroYak_CAF1 40020 112 - 1 UGAAUCUUUUGUCCACGUUCUGGACACCCGACUUCUAAUCCUCUGCCAAGUCCAGUGCCAAUGUAACGGCAGUUAAAGGGUCUUUAGUCAAAUUCUAUUAAUUAUUUGUUGA .((((....((((((.....))))))...((((....((((((((((.....((.......))....)))))....)))))....))))..))))................. ( -23.00) >consensus CGAAUCUUCCGUCCAGUUUGCGGACGGGCGACUUG_AUUCCUCGACCAAGUCUGGUGCCGAUGUAACGGCAGAUAAAUGAUCUUUAGUUAAACUUUAUUAAAUAUUUGUUGG ........((((((.......))))))..((((((...........))))))...(((((......)))))..(((..(((.((((((........)))))).)))..))). (-21.17 = -21.05 + -0.12)


| Location | 16,421,919 – 16,422,030 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 84.90 |
| Mean single sequence MFE | -41.92 |
| Consensus MFE | -31.20 |
| Energy contribution | -30.08 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16421919 111 + 22224390 CCGCCGUUACAUCGGCAACAAACUUGGUCGAGGAAU-CAAGUCGCCCGUCCGCAAACUGGACGGAAGAUUUGCCACCUUGAGGUGGAAUCACCGGCAACCGGCGUGAUCGCU ..((.(((((..((((((.....)))((((..((..-((((((..(((((((.....)))))))..))))))(((((....)))))..))..))))..)))..))))).)). ( -44.00) >DroSec_CAF1 42630 111 + 1 CUGCCGUUACAUCGGCAUCAGACUUGGUCGAGGAAC-CAAGUCGCCCGUCCGCAAACUGGACGGAACAUUCGCCACCUUGAGGUGAAACCACCGCCAACCGGCGUGAUCGCU ..((.(((((...(((....((((((((......))-))))))..(((((((.....))))))).......))).......((((....))))(((....)))))))).)). ( -45.40) >DroSim_CAF1 38906 111 + 1 CUGCCGUUACAUCGGCACCAGACUUGGUCGAGGAAC-CAAGUCGCCCGUCCGCAAACUGGACGGAACAUUCGCCACCUUGAGGUGGAACCACCGCCAACCGGCGUGAUCGCU ..((.(((((...(((....((((((((......))-))))))..(((((((.....))))))).......))).......((((....))))(((....)))))))).)). ( -46.80) >DroYak_CAF1 40060 112 + 1 CUGCCGUUACAUUGGCACUGGACUUGGCAGAGGAUUAGAAGUCGGGUGUCCAGAACGUGGACAAAAGAUUCAUUACAUUGAGGUGGAACAACCGGCAACCGGCGUGACCGCU ..((.(((((.((((..((((.(((....)))..))))..(((((.((((((.....))))))...(.((((((.......)))))).)..)))))..)))).))))).)). ( -31.50) >consensus CUGCCGUUACAUCGGCACCAGACUUGGUCGAGGAAC_CAAGUCGCCCGUCCGCAAACUGGACGGAACAUUCGCCACCUUGAGGUGGAACCACCGCCAACCGGCGUGAUCGCU ..((.(((((..(((.....((((((((......)).))))))..(((((((.....)))))))....((((((.......))))))...........)))..))))).)). (-31.20 = -30.08 + -1.13)

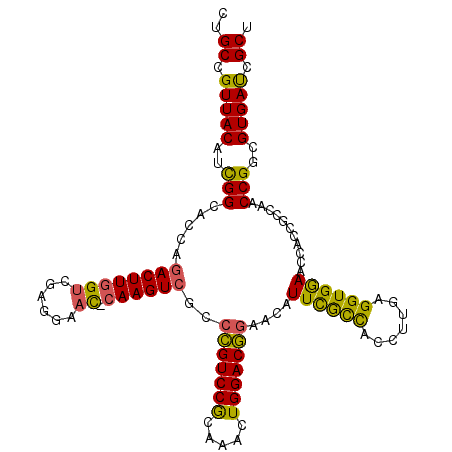

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:17 2006