| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,419,513 – 16,419,626 |
| Length | 113 |
| Max. P | 0.882383 |

| Location | 16,419,513 – 16,419,626 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -20.45 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16419513 113 + 22224390 AAAUUGGACAAACAAAUUUCAAAGAAACGCACAGAAAGAAACACAGUUUUUAUGAAACAAAUUGAUGUUUCAUAGAUAAAUAGUUUCUUAUCGUUUGGCAUAUAGAAUAUGCG ........(((((...((((...))))......(((((((((......(((((((((((......)))))))))))......))))))).)))))))((((((...)))))). ( -26.50) >DroSec_CAF1 40150 113 + 1 AAAUUGGACAAACAAAUUUCAAAGAAACGCACAGAAAGAAACACAGUUUUUAUGAAACAAAUUCAUGUUUCAUAGAUAAAUAGUUUCUUAUCGUUUUUCAUGUAGAAUAUUCG ......((.((((...((((...))))......(((((((((......(((((((((((......)))))))))))......))))))).)))))).)).............. ( -20.50) >DroSim_CAF1 36443 113 + 1 AAAUUGGACAAACAAAUUUCAAAGAAACGCACAGAAAGAAACAUAGUUUUUAUGAAGCAAACUGAUGUUUCAUAGAUAAAUAGUUUCUUAUCGUUUGGCAUGUAGAAUAUUCG ........(((((...((((...))))......(((((((((.((...(((((((((((......)))))))))))....))))))))).)))))))................ ( -22.00) >consensus AAAUUGGACAAACAAAUUUCAAAGAAACGCACAGAAAGAAACACAGUUUUUAUGAAACAAAUUGAUGUUUCAUAGAUAAAUAGUUUCUUAUCGUUUGGCAUGUAGAAUAUUCG ......(.(((((...((((...))))......(((((((((......(((((((((((......)))))))))))......))))))).))))))).).............. (-20.45 = -20.57 + 0.11)

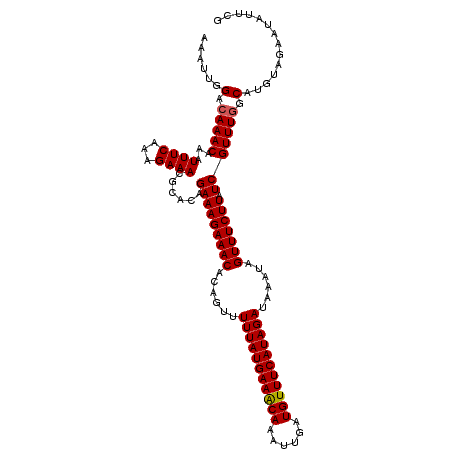
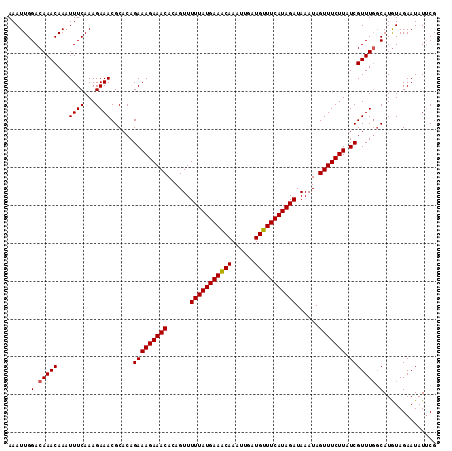
| Location | 16,419,513 – 16,419,626 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -18.28 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16419513 113 - 22224390 CGCAUAUUCUAUAUGCCAAACGAUAAGAAACUAUUUAUCUAUGAAACAUCAAUUUGUUUCAUAAAAACUGUGUUUCUUUCUGUGCGUUUCUUUGAAAUUUGUUUGUCCAAUUU .((((((...))))))(((((((.(((((((........(((((((((......)))))))))......(..(........)..))))))))......)))))))........ ( -24.70) >DroSec_CAF1 40150 113 - 1 CGAAUAUUCUACAUGAAAAACGAUAAGAAACUAUUUAUCUAUGAAACAUGAAUUUGUUUCAUAAAAACUGUGUUUCUUUCUGUGCGUUUCUUUGAAAUUUGUUUGUCCAAUUU ((((((.....((.((((.(((..(((((((........(((((((((......)))))))))........)))))))..)))...))))..)).....))))))........ ( -23.39) >DroSim_CAF1 36443 113 - 1 CGAAUAUUCUACAUGCCAAACGAUAAGAAACUAUUUAUCUAUGAAACAUCAGUUUGCUUCAUAAAAACUAUGUUUCUUUCUGUGCGUUUCUUUGAAAUUUGUUUGUCCAAUUU ................(((((((.((((((((((........(((((((.(((((.........)))))))))))).....))).)))))))......)))))))........ ( -19.72) >consensus CGAAUAUUCUACAUGCCAAACGAUAAGAAACUAUUUAUCUAUGAAACAUCAAUUUGUUUCAUAAAAACUGUGUUUCUUUCUGUGCGUUUCUUUGAAAUUUGUUUGUCCAAUUU ((((((.....((....(((((..(((((((........(((((((((......)))))))))........)))))))......)))))...)).....))))))........ (-18.28 = -18.72 + 0.45)

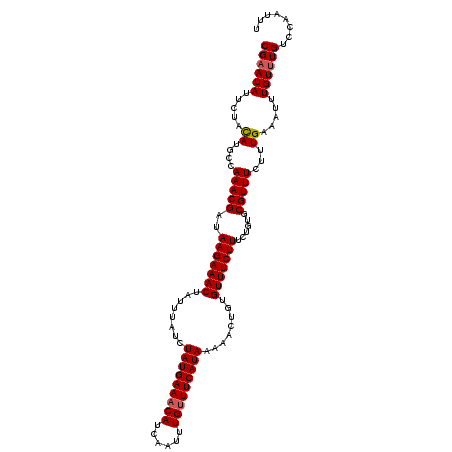

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:12 2006