
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,415,729 – 16,415,899 |
| Length | 170 |
| Max. P | 0.921226 |

| Location | 16,415,729 – 16,415,839 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.24 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -21.79 |
| Energy contribution | -22.23 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.664426 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16415729 110 - 22224390 GUUACCCCUAUGACAGAUAAACAGCUGAAUCUUCAGCUGAUUUAC---------UUA-AACUGAAAACUCUAUUACAAAAUAGUUGGGCAGCUGUGGUAGUUGAACCCGGACCACUGUUA ..........((((((.(((((((((((....))))))).)))).---------...-..(((.....((.(((((...((((((....)))))).))))).))...)))....)))))) ( -26.10) >DroSec_CAF1 36352 116 - 1 GUUACAGAUAUG---CAUAAACAGCUGAAUCUUCAGCUUAUUUACAUAAUUUACUUA-AACAGAAAACACUAUUACAAAAUAGUUGGGCAGCAGUGGUAGUUGGACCCAGACCACUGUUA ...........(---(......((((((....))))))...................-.........(((((((....))))).)).))(((((((((...((....)).))))))))). ( -25.50) >DroSim_CAF1 32659 117 - 1 GUUACAGAUAUG---CAUAAACAGCUGAAUCUUCAGCUGAUUUACUUAAUUUACUUAAAACAGAAAACACUAUUACAAAAUAGUUGGGCAGCAGUGGUAGUUGAACCCGGACCACUGUUU ..........((---(.(((((((((((....))))))).)))).......................(((((((....))))).)).)))((((((((...((....)).)))))))).. ( -27.20) >consensus GUUACAGAUAUG___CAUAAACAGCUGAAUCUUCAGCUGAUUUAC_UAAUUUACUUA_AACAGAAAACACUAUUACAAAAUAGUUGGGCAGCAGUGGUAGUUGAACCCGGACCACUGUUA (((..............(((((((((((....))))))).)))).........................(((((....)))))...)))(((((((((...((....)).))))))))). (-21.79 = -22.23 + 0.45)


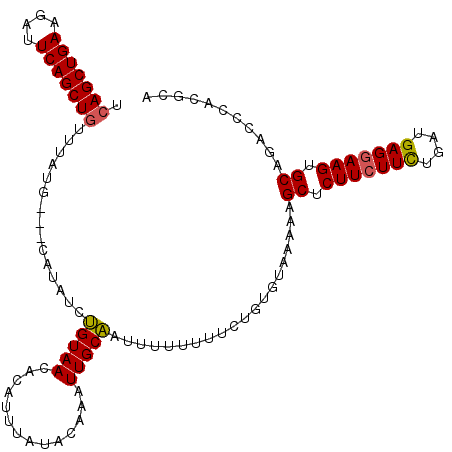
| Location | 16,415,799 – 16,415,899 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.14 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921226 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16415799 100 + 22224390 UCAGCUGAAGAUUCAGCUGUUUAUCUGUCAUAGGGGUAACAAUUUUUU-----UUGCCAAUUUUUUCCUGUGCAAAAAGCUCUUCUUUUGAUGAGGAAGUGCAGA-------- .(((((((....)))))))......((.(((((((((((.........-----)))))........))))))))....((.(((((((....))))))).))...-------- ( -28.60) >DroSec_CAF1 36431 110 + 1 UAAGCUGAAGAUUCAGCUGUUUAUG---CAUAUCUGUAACACAUUUAUACAAAUUGCAAUUUUUUUUCUGUGUAAAAAGCUCUUCUUCUGAUGAGAAAGUGCAGACCCACGCA ..((((((....)))))).....((---(...((((((.....((((((((.................))))))))...(((.((....)).)))....)))))).....))) ( -21.93) >DroSim_CAF1 32739 110 + 1 UCAGCUGAAGAUUCAGCUGUUUAUG---CAUAUCUGUAACACAUUUAUACAAAUUGCAAUUUUUUUUCUGUGUAAAAAGCUCUUCUUCUGAUGAGGAAGUGCAGACCCACGCA .(((((((....)))))))....((---(..............((((((((.................))))))))..((.(((((((....))))))).))........))) ( -28.63) >consensus UCAGCUGAAGAUUCAGCUGUUUAUG___CAUAUCUGUAACACAUUUAUACAAAUUGCAAUUUUUUUUCUGUGUAAAAAGCUCUUCUUCUGAUGAGGAAGUGCAGACCCACGCA .(((((((....)))))))...............(((((..............)))))....................((.(((((((....))))))).))........... (-19.67 = -19.67 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:03 2006