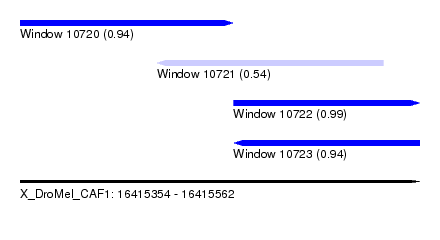
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,415,354 – 16,415,562 |
| Length | 208 |
| Max. P | 0.986187 |

| Location | 16,415,354 – 16,415,465 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -23.59 |
| Consensus MFE | -20.36 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.935645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16415354 111 + 22224390 AAAAUUUGAAUUGCUACCCGAAAGUUGAAUUUUAAUUUGGGCACUUCAUAAAGGCCAACGUGAGUUUCCUUUUAUGUUUAUUUUGGUUAGUGCCUUUUCCUCUUCCGACUU .....................((((((...........(((((((.((((((((..(((....)))..))))))))............)))))))..........)))))) ( -23.82) >DroSec_CAF1 35978 111 + 1 AAAAUUCGAAUUGCUACCCGAAAGUUGAAUUUUAAUUUAGGCACUUCAUAAAGGCCAACGUGAGUUUCCUUUUAUGUUUAUUUUGGUUAGUGCCUUUUCCUCUUCCGACUU (((((((((....(.....)....))))))))).....(((((((.((((((((..(((....)))..))))))))............)))))))................ ( -24.72) >DroSim_CAF1 32282 111 + 1 AAAAUUUGAAUUGCUACCCGAAAGUUGAAUUUUAAUUUGGGCACUUCAUAAAGGCCAACGUGAGUUCCCUUUUAUGUUUAUUUUGGUUAGUGCCUUUUCCUCUUCCGACUU .....................((((((...........(((((((.((((((((..(((....)))..))))))))............)))))))..........)))))) ( -23.82) >DroYak_CAF1 33893 97 + 1 --------------UACCCAAAUUUUUCAUUUUAACUUGGGCACUUCAUAAAGGCCAACGUGAGUUUCCUUUUAUGUUUAUUUUGGUUAGUGCCUUUUCCUGUGCCGACUU --------------.........................(((((..((((((((..(((....)))..))))))))........(((....))).......)))))..... ( -22.00) >consensus AAAAUUUGAAUUGCUACCCGAAAGUUGAAUUUUAAUUUGGGCACUUCAUAAAGGCCAACGUGAGUUUCCUUUUAUGUUUAUUUUGGUUAGUGCCUUUUCCUCUUCCGACUU .....................((((((...........(((((((...(((((...((((((((......))))))))..)))))...)))))))..........)))))) (-20.36 = -20.93 + 0.56)


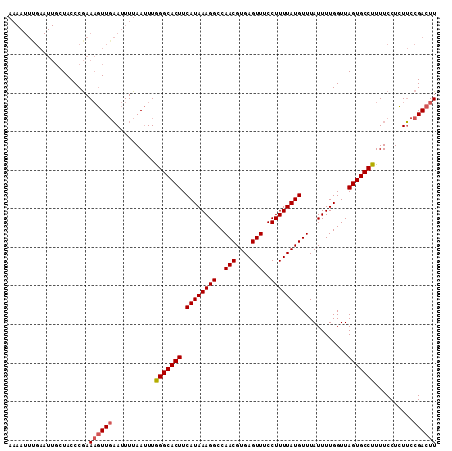
| Location | 16,415,425 – 16,415,543 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -27.48 |
| Energy contribution | -27.35 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16415425 118 - 22224390 UUUCUGCGCUUCAUCCUUCGCAUCCUCUUCUGCUGCAGAUGACGUUUCGGGCCCAAGAAAAAGGAUGCGUGCUGAUGAAAGUCGGAAGAGGAAAAGGCACUAACCAAAAUAAACAUAA .......((((..(((((((((((((.((((...((.((.......))..))...))))..))))))))..(((((....)))))..)))))..)))).................... ( -34.10) >DroSec_CAF1 36049 115 - 1 UUUCUG--UUUCAUCCUUCGCAUCCUCUUCUGCUGCAGAUGACGUUUCG-GCCCAAGAAAAAGGAUGCGUGCUGAUGAAAGUCGGAAGAGGAAAAGGCACUAACCAAAAUAAACAUAA ....((--(((..(((((((((((((.((((...((.((.......)).-))...))))..))))))))..(((((....)))))..)))))..)))))................... ( -31.80) >DroSim_CAF1 32353 116 - 1 UUUCUG--CUUCAUCCUUCGCAUCCUCUUCUGCUGCAGAUGACGUUUCGGGUCCAAGAAAAAGGAUGCGUGCUGAUGAAAGUCGGAAGAGGAAAAGGCACUAACCAAAAUAAACAUAA ....((--(((..(((((((((((((.((((...((.((.......))..))...))))..))))))))..(((((....)))))..)))))..)))))................... ( -32.70) >DroYak_CAF1 33950 116 - 1 UUUCUG--CUUCAUCCUUCGCAUCCUUUUUUGCUGCAGAUGACGUUUCGGGCCCAAGAAAAAGGAUGCGUGCUGAUGAAAGUCGGCACAGGAAAAGGCACUAACCAAAAUAAACAUAA ....((--(((..((((..((((((((((((...((.((.......))..))....))))))))))))((((((((....))))))))))))..)))))................... ( -42.90) >consensus UUUCUG__CUUCAUCCUUCGCAUCCUCUUCUGCUGCAGAUGACGUUUCGGGCCCAAGAAAAAGGAUGCGUGCUGAUGAAAGUCGGAAGAGGAAAAGGCACUAACCAAAAUAAACAUAA .............(((((((((((((.((((...((...(((....))).))...))))..))))))))..(((((....)))))..)))))...((......))............. (-27.48 = -27.35 + -0.13)

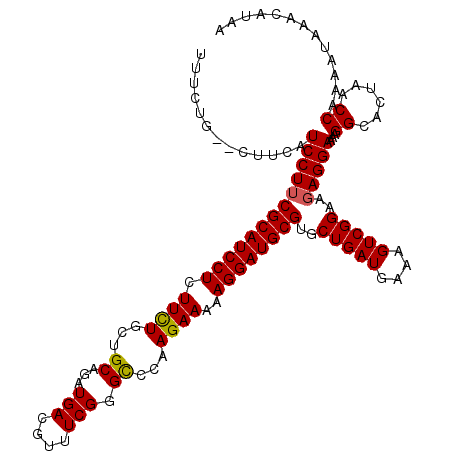

| Location | 16,415,465 – 16,415,562 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -27.94 |
| Energy contribution | -27.38 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16415465 97 + 22224390 UCAUCAGCACGCAUCCUUUUUCUUGGGCCCGAAACGUCAUCUGCAGCAGAAGAGGAUGCGAAGGAUGAAGCGCAGAAACGGCAACGAAUGCCAAGUA (((((....(((((((((((.((...((..((.......)).))...)))))))))))))...))))).(.(((....((....))..))))..... ( -29.40) >DroSec_CAF1 36089 94 + 1 UCAUCAGCACGCAUCCUUUUUCUUGGGC-CGAAACGUCAUCUGCAGCAGAAGAGGAUGCGAAGGAUGAAA--CAGAAACGGCAAAGAAUGCCAAGUA (((((....(((((((((((.((...((-.((.......)).))...)))))))))))))...)))))..--.......((((.....))))..... ( -30.70) >DroSim_CAF1 32393 95 + 1 UCAUCAGCACGCAUCCUUUUUCUUGGACCCGAAACGUCAUCUGCAGCAGAAGAGGAUGCGAAGGAUGAAG--CAGAAACGGCAAAGAAUGCCAAGUA (((((....((((((((((......(((.......))).((((...))))))))))))))...)))))..--.......((((.....))))..... ( -29.00) >DroYak_CAF1 33990 95 + 1 UCAUCAGCACGCAUCCUUUUUCUUGGGCCCGAAACGUCAUCUGCAGCAAAAAAGGAUGCGAAGGAUGAAG--CAGAAACGGCAAAAAAUGCCAAGUA (((((....((((((((((((..((.((..((.......)).))..))))))))))))))...)))))..--.......((((.....))))..... ( -28.10) >consensus UCAUCAGCACGCAUCCUUUUUCUUGGGCCCGAAACGUCAUCUGCAGCAGAAGAGGAUGCGAAGGAUGAAG__CAGAAACGGCAAAGAAUGCCAAGUA (((((....((((((((((((.(((.(...((....))...).)))..))))))))))))...)))))...........((((.....))))..... (-27.94 = -27.38 + -0.56)



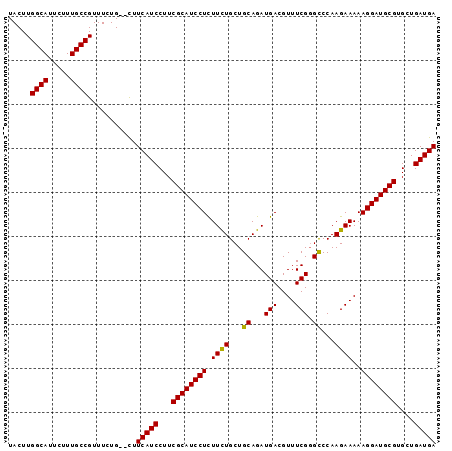
| Location | 16,415,465 – 16,415,562 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -27.62 |
| Energy contribution | -27.25 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16415465 97 - 22224390 UACUUGGCAUUCGUUGCCGUUUCUGCGCUUCAUCCUUCGCAUCCUCUUCUGCUGCAGAUGACGUUUCGGGCCCAAGAAAAAGGAUGCGUGCUGAUGA .....((((.....))))...........(((((...((((((((.((((...((.((.......))..))...))))..))))))))....))))) ( -29.10) >DroSec_CAF1 36089 94 - 1 UACUUGGCAUUCUUUGCCGUUUCUG--UUUCAUCCUUCGCAUCCUCUUCUGCUGCAGAUGACGUUUCG-GCCCAAGAAAAAGGAUGCGUGCUGAUGA .....((((.....)))).......--..(((((...((((((((.((((...((.((.......)).-))...))))..))))))))....))))) ( -29.10) >DroSim_CAF1 32393 95 - 1 UACUUGGCAUUCUUUGCCGUUUCUG--CUUCAUCCUUCGCAUCCUCUUCUGCUGCAGAUGACGUUUCGGGUCCAAGAAAAAGGAUGCGUGCUGAUGA .....((((.....)))).......--..(((((...((((((((.((((...((.((.......))..))...))))..))))))))....))))) ( -27.50) >DroYak_CAF1 33990 95 - 1 UACUUGGCAUUUUUUGCCGUUUCUG--CUUCAUCCUUCGCAUCCUUUUUUGCUGCAGAUGACGUUUCGGGCCCAAGAAAAAGGAUGCGUGCUGAUGA .....((((.....)))).......--..(((((...(((((((((((((...((.((.......))..))....)))))))))))))....))))) ( -31.70) >consensus UACUUGGCAUUCUUUGCCGUUUCUG__CUUCAUCCUUCGCAUCCUCUUCUGCUGCAGAUGACGUUUCGGGCCCAAGAAAAAGGAUGCGUGCUGAUGA .....((((.....))))...........(((((...((((((((.((((...((...(((....))).))...))))..))))))))....))))) (-27.62 = -27.25 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:17:02 2006