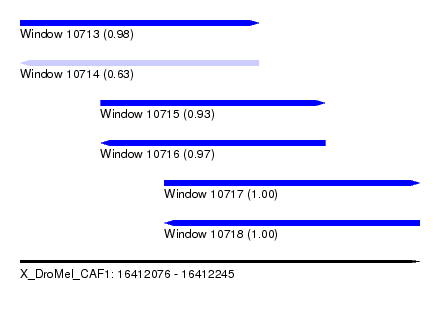
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,412,076 – 16,412,245 |
| Length | 169 |
| Max. P | 0.999875 |

| Location | 16,412,076 – 16,412,177 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16412076 101 + 22224390 UUUUCCUCUGCUUUCUUUCUCAGACAU---UUUUUUU---UUUUUUGGUUAGUUGCCCGCUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAA ......((((..........))))...---...((((---(.(((((.((((((((.(((.....)))((((......))))..))))))))))))).))))).... ( -19.50) >DroSec_CAF1 32735 104 + 1 UUUUCCUCUGCUUUAGUUCUCAGACAUCUCUUUUUUU---UUUUUUUGUUAGUUGCCCGUUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAA ......((((..........))))..........(((---((.(((((((((((((....((((((((........))))))))))))))))))))).))))).... ( -22.60) >DroSim_CAF1 28946 107 + 1 UUUUCCUCUGCUUUCUUUAUCAGACAUCUCUUUUUUUUUUUUUUUUUGUUAGUUGCCCGUUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAA ......((((.(......).)))).............(((((.(((((((((((((....((((((((........))))))))))))))))))))).))))).... ( -22.90) >DroYak_CAF1 30794 95 + 1 UUCUCCUCUAUUUUCUUUUGCAGU---------UCUU---UUUUUUUUUUAGUUGCCCGCUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACACA ..........((((.(((((((((---------(...---...........(.....)((((((((((........)))))))))))))).)))))).))))..... ( -17.40) >consensus UUUUCCUCUGCUUUCUUUCUCAGACAU___UUUUUUU___UUUUUUUGUUAGUUGCCCGCUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAA ...........................................(((((((((((((....((((((((........))))))))))))))))))))).......... (-17.60 = -18.10 + 0.50)



| Location | 16,412,076 – 16,412,177 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.14 |
| Mean single sequence MFE | -18.52 |
| Consensus MFE | -13.90 |
| Energy contribution | -13.78 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16412076 101 - 22224390 UUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAGCGGGCAACUAACCAAAAAA---AAAAAAA---AUGUCUGAGAAAGAAAGCAGAGGAAAA .....((((.(((((((..((((((((((......))))......))))))................---.......---...(((.....))).))))))).)))) ( -18.90) >DroSec_CAF1 32735 104 - 1 UUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAACGGGCAACUAACAAAAAAA---AAAAAAAGAGAUGUCUGAGAACUAAAGCAGAGGAAAA ((((((((.(((((((.....))..((((......)))).........((((((.((..........---........)).))))))))))).))))))))...... ( -18.87) >DroSim_CAF1 28946 107 - 1 UUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAACGGGCAACUAACAAAAAAAAAAAAAAAAAGAGAUGUCUGAUAAAGAAAGCAGAGGAAAA .....((((.(((((((..((....((((......)))).........((((((.((.....................)).))))))....))..))))))).)))) ( -19.20) >DroYak_CAF1 30794 95 - 1 UGUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAGCGGGCAACUAAAAAAAAAA---AAGA---------ACUGCAAAAGAAAAUAGAGGAGAA (.(((((((.((((((...((((((((((......))))......))))((....))..........---.)).---------...))))))))))))).)...... ( -17.10) >consensus UUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAACGGGCAACUAACAAAAAAA___AAAAAAA___AUGUCUGAGAAAGAAAGCAGAGGAAAA ((((((((((((((((.....))..((((......)))).....)))))((....))...................................)))))))))...... (-13.90 = -13.78 + -0.12)



| Location | 16,412,110 – 16,412,205 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -21.81 |
| Consensus MFE | -20.22 |
| Energy contribution | -19.96 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16412110 95 + 22224390 UUUUUUGGUUAGUUGCCCGCUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAAGCCAGCGAACAAAACCAAAGCAACCGCC ..((((((((..((((..(((((((.((((((......)))...((.....))...)))...)))))))..))))....))))))))........ ( -25.70) >DroSec_CAF1 32772 95 + 1 UUUUUUUGUUAGUUGCCCGUUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAAGCCAGCCAACAAAACCAAAGCAACCGCC ...(((((((((((((....((((((((........))))))))))))))))))))).............................((....)). ( -20.30) >DroSim_CAF1 28986 95 + 1 UUUUUUUGUUAGUUGCCCGUUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAAGCCAGCCAACAAAACCAAAGCAACCGCC ...(((((((((((((....((((((((........))))))))))))))))))))).............................((....)). ( -20.30) >DroYak_CAF1 30822 95 + 1 UUUUUUUUUUAGUUGCCCGCUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACACAGCCAGCCAACAAAACCAAAGCAACCGCC ...........(((((..((((((((((........)))))))))).(((.((..............)).))).............))))).... ( -20.94) >consensus UUUUUUUGUUAGUUGCCCGCUUUGUGUGUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAAGCCAGCCAACAAAACCAAAGCAACCGCC ...........(((((..((((((((((........)))))))))).(((.((..............)).))).............))))).... (-20.22 = -19.96 + -0.25)



| Location | 16,412,110 – 16,412,205 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 97.19 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -27.39 |
| Energy contribution | -27.39 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16412110 95 - 22224390 GGCGGUUGCUUUGGUUUUGUUCGCUGGCUUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAGCGGGCAACUAACCAAAAAA ((.((((((((..(((((((..((((((..............)))))).....((((......))))...)))))))))))))))..))...... ( -28.54) >DroSec_CAF1 32772 95 - 1 GGCGGUUGCUUUGGUUUUGUUGGCUGGCUUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAACGGGCAACUAACAAAAAAA (..((((((((..(((((((.(((((((..............)))))))....((((......))))...)))))))))))))))..)....... ( -29.24) >DroSim_CAF1 28986 95 - 1 GGCGGUUGCUUUGGUUUUGUUGGCUGGCUUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAACGGGCAACUAACAAAAAAA (..((((((((..(((((((.(((((((..............)))))))....((((......))))...)))))))))))))))..)....... ( -29.24) >DroYak_CAF1 30822 95 - 1 GGCGGUUGCUUUGGUUUUGUUGGCUGGCUGUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAGCGGGCAACUAAAAAAAAAA ...((((((((..(((((((.(((((((..............)))))))....((((......))))...))))))))))))))).......... ( -28.14) >consensus GGCGGUUGCUUUGGUUUUGUUGGCUGGCUUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCACACACAAAACGGGCAACUAACAAAAAAA ...((((((((..(((((((.(((((((..............)))))))....((((......))))...))))))))))))))).......... (-27.39 = -27.39 + 0.00)

| Location | 16,412,137 – 16,412,245 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -32.04 |
| Consensus MFE | -28.03 |
| Energy contribution | -28.07 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16412137 108 + 22224390 GUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAA-----GCCAGCGAACAAAACCAAAGCAACC-------GCCUUUGUUGUUGGCUAACGGCAUUGUUUGCAUGCUGAGUGCA .((((......))))..(((..(((((......((((((.-----((((((.(((((...((((((....-------).)))))))))).)))...))).))))))...)))))..))). ( -32.20) >DroSec_CAF1 32799 108 + 1 GUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAA-----GCCAGCCAACAAAACCAAAGCAACC-------GCCUUUGUUGCUGGCUAACGGCAUUGUUUGCAUGCUGAGUGCA ...........((((...((((..(((((.(((...(..(-----((((((.((((((......((....-------)).)))))))))))))...)...))))))))..))))..)))) ( -32.50) >DroSim_CAF1 29013 108 + 1 GUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAA-----GCCAGCCAACAAAACCAAAGCAACC-------GCCUUUGUUGCUGGCUAACGGCAUUGUUUGCAUGCUGAGUGCA ...........((((...((((..(((((.(((...(..(-----((((((.((((((......((....-------)).)))))))))))))...)...))))))))..))))..)))) ( -32.50) >DroYak_CAF1 30849 108 + 1 GUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACACA-----GCCAGCCAACAAAACCAAAGCAACC-------GCCUUUGUUGCUGGCUAACGGCAUUGUUUGCAUGCUGAGUGCA ...........((((...((((..(((((.(((...(..(-----((((((.((((((......((....-------)).)))))))))))))...)...))))))))..))))..)))) ( -32.50) >DroAna_CAF1 34637 120 + 1 GUGCAAAUUUAUGCAAAGCAGCUAGCCAAACAGAAACAAAGCGAAGCCAGCAAACAAAACCAAAGCAACCGAAAGCAGCUUUUGUUGCUCGCUAACGGCAUUGUUUGCAUGCCGAGUGCA .((((......))))..(((.((................(((((.((.((((((.........(((...(....)..))))))))))))))))..((((((.(....)))))))))))). ( -30.50) >consensus GUGCAAAUUUAUGCAAAGCAGCUAGCAAAACAAAAACAAA_____GCCAGCCAACAAAACCAAAGCAACC_______GCCUUUGUUGCUGGCUAACGGCAUUGUUUGCAUGCUGAGUGCA ((((((((..((((...((.....))...................((((((.((((((......................)))))))))))).....)))).))))))))((.....)). (-28.03 = -28.07 + 0.04)


| Location | 16,412,137 – 16,412,245 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -37.76 |
| Consensus MFE | -32.30 |
| Energy contribution | -32.18 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16412137 108 - 22224390 UGCACUCAGCAUGCAAACAAUGCCGUUAGCCAACAACAAAGGC-------GGUUGCUUUGGUUUUGUUCGCUGGC-----UUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCAC ((((......(((((((....((.(((((((((.(((((((.(-------(((.((.........))..)))).)-----)))))).)))....)))))).))))))))).....)))). ( -32.20) >DroSec_CAF1 32799 108 - 1 UGCACUCAGCAUGCAAACAAUGCCGUUAGCCAGCAACAAAGGC-------GGUUGCUUUGGUUUUGUUGGCUGGC-----UUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCAC .(((((.((((.((((((((.(((...(((((((((..(((((-------....)))))....))))))))))))-----.))))..))))..)))))).)))..((((......)))). ( -38.10) >DroSim_CAF1 29013 108 - 1 UGCACUCAGCAUGCAAACAAUGCCGUUAGCCAGCAACAAAGGC-------GGUUGCUUUGGUUUUGUUGGCUGGC-----UUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCAC .(((((.((((.((((((((.(((...(((((((((..(((((-------....)))))....))))))))))))-----.))))..))))..)))))).)))..((((......)))). ( -38.10) >DroYak_CAF1 30849 108 - 1 UGCACUCAGCAUGCAAACAAUGCCGUUAGCCAGCAACAAAGGC-------GGUUGCUUUGGUUUUGUUGGCUGGC-----UGUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCAC .(((((.((((.(((((.(((((.((((((((((((..(((((-------....)))))....))))))))))))-----.))))))))))..)))))).)))..((((......)))). ( -40.90) >DroAna_CAF1 34637 120 - 1 UGCACUCGGCAUGCAAACAAUGCCGUUAGCGAGCAACAAAAGCUGCUUUCGGUUGCUUUGGUUUUGUUUGCUGGCUUCGCUUUGUUUCUGUUUGGCUAGCUGCUUUGCAUAAAUUUGCAC .......(((((.......)))))((((((((((((..(((((.((.....)).)))))....))))))))))))...((...((((.(((..(((.....)))..))).))))..)).. ( -39.50) >consensus UGCACUCAGCAUGCAAACAAUGCCGUUAGCCAGCAACAAAGGC_______GGUUGCUUUGGUUUUGUUGGCUGGC_____UUUGUUUUUGUUUUGCUAGCUGCUUUGCAUAAAUUUGCAC .(((((.((((.((((((((....((((((((((((..(((((...........)))))....))))))))))))......)))))..)))..)))))).)))..((((......)))). (-32.30 = -32.18 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:57 2006