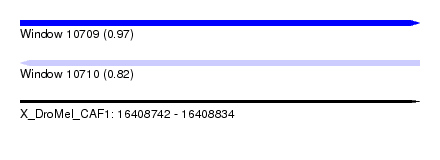
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,408,742 – 16,408,834 |
| Length | 92 |
| Max. P | 0.965818 |

| Location | 16,408,742 – 16,408,834 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.36 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
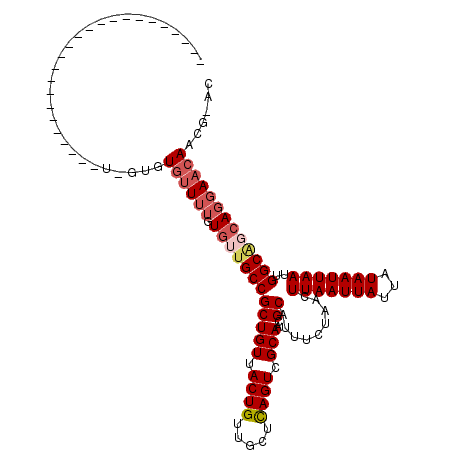
>X_DroMel_CAF1 16408742 92 + 22224390 GUUCGUUGUUCCUGAUGCCAAUUAAUUAUAAUAAUUAAGUUAGAAAUGCGUUGCGACUGAGCAACAGUAACAGCGGCAACACAAAACACAC-G------------------------ ....(((((..(((.(((...(((((((...)))))))...........(((((......))))).))).)))..)))))...........-.------------------------ ( -20.80) >DroSec_CAF1 29485 91 + 1 GU-CGUUGUUCCUGCUGCCAAUUAAUUAUAAUAAUUAAGUUAGAAAUGCGUUGCGACUGAGCAACAGUAACAGCGGCAACACAAAACACAC-A------------------------ ((-(((((((.((((......(((((((...))))))).........))(((((......))))))).)))))))))..............-.------------------------ ( -22.66) >DroSim_CAF1 26413 92 + 1 GU-CGUUGUUCCUGCUGCCAAUUAAUUAUAAUAAUUAAGUUAGAAAUGCGUUGCGACUGAGCAACAGUAACAGCGGCAACACAAAACACACCA------------------------ ((-(((((((.((((......(((((((...))))))).........))(((((......))))))).)))))))))................------------------------ ( -22.66) >DroYak_CAF1 27507 89 + 1 GU-GGUUGUUCCUGCCGCCAAUUAAUUAUAAUAAUUAAGUUAGAAAUGCGUUGCGACUGAGCAACAGUAACAGCGGCAACACACAACA--G-G------------------------ ..-.(((((...((((((...(((((((...)))))))........((((((((......))))).)))...))))))....))))).--.-.------------------------ ( -24.90) >DroAna_CAF1 30630 113 + 1 UU-AGUUGUUCUU---GCCAAUUAAUUAUAAUAAUUAAGUUCGAAAUGCGUUGCGACUAAGCAACAGUAACAGCGGCAACAACAAUUAUGCCAUCAAUGGAAAGCGGCCAGGAACCA ..-....((((((---(((..(((((((...))))))).........(((((((......))))).........((((..........))))...........)))).))))))).. ( -26.60) >consensus GU_CGUUGUUCCUGCUGCCAAUUAAUUAUAAUAAUUAAGUUAGAAAUGCGUUGCGACUGAGCAACAGUAACAGCGGCAACACAAAACACAC_A________________________ ....(((((..(((.(((...(((((((...)))))))...........(((((......))))).))).)))..)))))..................................... (-18.92 = -19.36 + 0.44)



| Location | 16,408,742 – 16,408,834 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.22 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.92 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16408742 92 - 22224390 ------------------------C-GUGUGUUUUGUGUUGCCGCUGUUACUGUUGCUCAGUCGCAACGCAUUUCUAACUUAAUUAUUAUAAUUAAUUGGCAUCAGGAACAACGAAC ------------------------(-((.(((((((((((((.((((...........)))).)))))))....((((.(((((((...))))))))))).....)))))))))... ( -22.40) >DroSec_CAF1 29485 91 - 1 ------------------------U-GUGUGUUUUGUGUUGCCGCUGUUACUGUUGCUCAGUCGCAACGCAUUUCUAACUUAAUUAUUAUAAUUAAUUGGCAGCAGGAACAACG-AC ------------------------.-((.((((((.((((((((((((.((((.....)))).)))..)).........(((((((...)))))))..))))))))))))))).-.. ( -23.60) >DroSim_CAF1 26413 92 - 1 ------------------------UGGUGUGUUUUGUGUUGCCGCUGUUACUGUUGCUCAGUCGCAACGCAUUUCUAACUUAAUUAUUAUAAUUAAUUGGCAGCAGGAACAACG-AC ------------------------..((.((((((.((((((((((((.((((.....)))).)))..)).........(((((((...)))))))..))))))))))))))).-.. ( -23.60) >DroYak_CAF1 27507 89 - 1 ------------------------C-C--UGUUGUGUGUUGCCGCUGUUACUGUUGCUCAGUCGCAACGCAUUUCUAACUUAAUUAUUAUAAUUAAUUGGCGGCAGGAACAACC-AC ------------------------(-(--(((((((((((((.((((...........)))).)))))))..........((((((.......)))))))))))))).......-.. ( -27.10) >DroAna_CAF1 30630 113 - 1 UGGUUCCUGGCCGCUUUCCAUUGAUGGCAUAAUUGUUGUUGCCGCUGUUACUGUUGCUUAGUCGCAACGCAUUUCGAACUUAAUUAUUAUAAUUAAUUGGC---AAGAACAACU-AA .(((.....)))(((.((....)).)))....(((((.(((((.........(((((......)))))...........(((((((...)))))))..)))---)).)))))..-.. ( -26.20) >consensus ________________________U_GUGUGUUUUGUGUUGCCGCUGUUACUGUUGCUCAGUCGCAACGCAUUUCUAACUUAAUUAUUAUAAUUAAUUGGCAGCAGGAACAACG_AC .............................((((((.((((((((((((.((((.....)))).)))..)).........(((((((...)))))))..)))))))))))))...... (-17.60 = -18.92 + 1.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:51 2006