
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,400,804 – 16,401,044 |
| Length | 240 |
| Max. P | 0.895504 |

| Location | 16,400,804 – 16,400,924 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -53.67 |
| Consensus MFE | -48.73 |
| Energy contribution | -49.57 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16400804 120 + 22224390 ACGUCGAGAUAGUUGGGCGCCGAGAAGAUGGUGAUCAGCGAGGGGAAGCCGGUGGUCUGGCUUUUGCGGUACAUGCGGUAGCCGGCGUCCUGCGCCUCGUGCGCCCGGAUGAUCGACAGC ..(((((.....((((((((((((..(((....))).(((..((((.(((((....))((((.(((((.....))))).))))))).)))).))))))).))))))))....)))))... ( -56.00) >DroGri_CAF1 20400 120 + 1 ACGUCCAGAUAAUUGGGCGCCGAGAAGAUGGUGAUCAGCGAGGGAAAACCGGUGGUCUGGCUCUUGCGGUACAUGCGGUAGCCGGCGUCCUGGGCCUCGUGCGCCCGUAUGAUCGACAGC ..(((..(((...(((((((......(((....))).((((((.....((((....((((((.(((((.....))))).))))))....)))).)))))))))))))....))))))... ( -46.30) >DroSec_CAF1 21717 120 + 1 ACGUCGAGAUAGUUGGGCGCCGAGAAAAUGGUGAUCAGCGAGGGGAAGCCGGUGGUCUGGCUCUUGCGGUACAUGCGGUAGCCGGCGUCCUGCGCCUCGUGCGCCCGGAUGAUCGACAGC ..(((((.....((((((((((((....((.....))(((..((((.(((((....))((((.(((((.....))))).))))))).)))).))))))).))))))))....)))))... ( -53.80) >DroYak_CAF1 18343 120 + 1 ACGUCGAGAUAGUUGGGCGCCGAGAAGAUGGUGAUCAGCGAGGGGAAGCCGGUGGUCUGGCUCUUGCGGUACAUGCGGUAGCCGGCGUCCUGCGCCUCGUGCGCCCGGAUGAUCGACAGC ..(((((.....((((((((((((..(((....))).(((..((((.(((((....))((((.(((((.....))))).))))))).)))).))))))).))))))))....)))))... ( -56.00) >DroMoj_CAF1 20078 120 + 1 ACGUCGAGAUAGUUGGGCGCCGAGAAGAUGGUGAUCAGCGAGGGGAAACCGGUGGUCUGGCUCUUGCGGUACAUGCGGUAGCCGGCGUCCUGCGCCUCGUGCGCCCGUAUGAUCGACAGC ..(((((......(((((((((((..(((....))).(((..((....))((..(.((((((.(((((.....))))).)))))))..))..))))))).))))))).....)))))... ( -53.80) >DroAna_CAF1 19170 120 + 1 ACGUCGAGAUAGUUGGGCGCCGAGAAGAUGGUGAUCAGCGAGGGGAAGCCGGUGGUCUGGCUCUUCCGGUACAUGCGGUAGCCGGCGUCCUGCGCCUCGUGCGCCCGGAUGAUCGACAGC ..(((((.....((((((((((((..(((....))).(((..((((.((((((.((((((.....)))).))........)))))).)))).))))))).))))))))....)))))... ( -56.10) >consensus ACGUCGAGAUAGUUGGGCGCCGAGAAGAUGGUGAUCAGCGAGGGGAAGCCGGUGGUCUGGCUCUUGCGGUACAUGCGGUAGCCGGCGUCCUGCGCCUCGUGCGCCCGGAUGAUCGACAGC ..(((((.....(((((((((((.........(((((.((.(......))).))))).((((.(((((.....))))).))))((((.....))))))).))))))))....)))))... (-48.73 = -49.57 + 0.83)

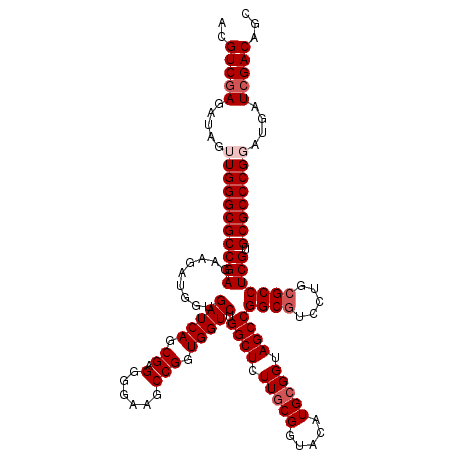
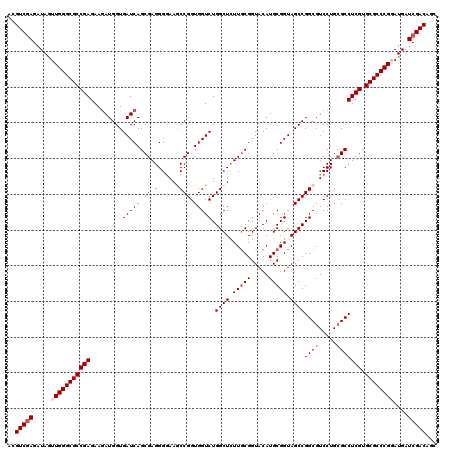
| Location | 16,400,804 – 16,400,924 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -42.85 |
| Consensus MFE | -41.07 |
| Energy contribution | -41.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16400804 120 - 22224390 GCUGUCGAUCAUCCGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAAAGCCAGACCACCGGCUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGU ...(((((......((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...))).(((....)))..)))))))))).......))))).. ( -43.62) >DroGri_CAF1 20400 120 - 1 GCUGUCGAUCAUACGGGCGCACGAGGCCCAGGACGCCGGCUACCGCAUGUACCGCAAGAGCCAGACCACCGGUUUUCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAAUUAUCUGGACGU ...((((((.((..((((((.((((((...(((.(((((((...((.......))...)))).(.....))))..)))...)).(((....)))..)))))))))).)).)))..))).. ( -35.40) >DroSec_CAF1 21717 120 - 1 GCUGUCGAUCAUCCGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAGAGCCAGACCACCGGCUUCCCCUCGCUGAUCACCAUUUUCUCGGCGCCCAACUAUCUCGACGU ...(((((......((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...)))((.....))....)))))))))).......))))).. ( -43.72) >DroYak_CAF1 18343 120 - 1 GCUGUCGAUCAUCCGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAGAGCCAGACCACCGGCUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGU ...(((((......((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...))).(((....)))..)))))))))).......))))).. ( -44.72) >DroMoj_CAF1 20078 120 - 1 GCUGUCGAUCAUACGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAGAGCCAGACCACCGGUUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGU ...(((((......((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...))).(((....)))..)))))))))).......))))).. ( -43.22) >DroAna_CAF1 19170 120 - 1 GCUGUCGAUCAUCCGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGGAAGAGCCAGACCACCGGCUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGU ...(((((......((((((.(((((((.(((...((((.(((.....)))))))..(((((........)))))..)))))).(((....)))..)))))))))).......))))).. ( -46.42) >consensus GCUGUCGAUCAUCCGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAGAGCCAGACCACCGGCUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGU ...(((((......((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...))).(((....)))..)))))))))).......))))).. (-41.07 = -41.07 + 0.00)

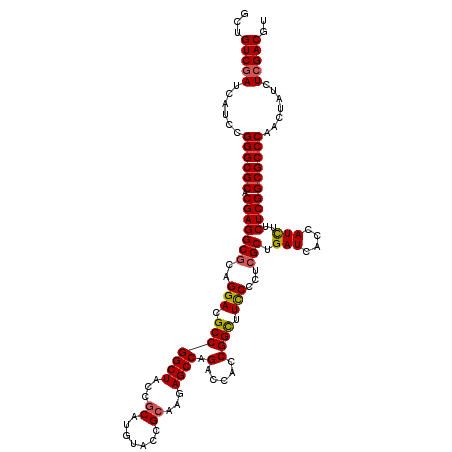
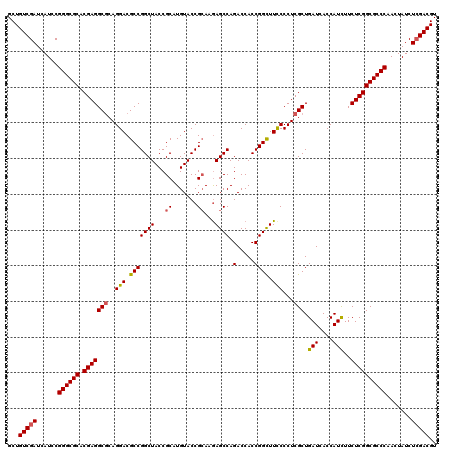
| Location | 16,400,924 – 16,401,044 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -35.18 |
| Energy contribution | -35.23 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16400924 120 - 22224390 UCCGCUGGAGGACUUCGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCCGUGCGCGGCUGCUCGUACUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCU ((.((.(((((....(((((.((((.....))))...........((((......))))(((((.(((.(((....))))))...))))).)))))...))))).)).)).......... ( -38.90) >DroGri_CAF1 20520 120 - 1 UCCGCUCGAGGACUUCGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCGGUGCGCGGCUGUUCGUAUUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCU ...(..((((..(((((....)))..))..))))..)........((((......))))(((((.(((.(((....))))))...)))))...(((((......)))))........... ( -35.10) >DroSec_CAF1 21837 120 - 1 UCCGCUGGAGGACUUCGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCCGUGCGCGGCUGCUCGUACUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCU ((.((.(((((....(((((.((((.....))))...........((((......))))(((((.(((.(((....))))))...))))).)))))...))))).)).)).......... ( -38.90) >DroYak_CAF1 18463 120 - 1 UCCGCUGGAGGACUUCGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCCGUGCGCGGCUGCUCGUACUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCU ((.((.(((((....(((((.((((.....))))...........((((......))))(((((.(((.(((....))))))...))))).)))))...))))).)).)).......... ( -38.90) >DroMoj_CAF1 20198 120 - 1 UCCGCUCGAGGACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCGGUGCGCGGCUGUUCGUAUUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAAUCU ...(..((((..(((((....)..))))..))))..)........((((......))))(((((.(((.(((....))))))...)))))...(((((......)))))........... ( -33.00) >DroAna_CAF1 19290 120 - 1 UCCGCUGGAGGACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCGGUGCGCGGGUGCUCGUACUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCU ....(((.((((...(.(((.((((.....))))...........((((......))))(((((((..((((((.....)).)))).)))))))))).)..)))).)))........... ( -36.70) >consensus UCCGCUGGAGGACUUCGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCCGUGCGCGGCUGCUCGUACUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCU ((.((.(((((....(((((.((((.....))))...........((((......))))(((((.(((.(((....))))))...))))).)))))...))))).)).)).......... (-35.18 = -35.23 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:43 2006