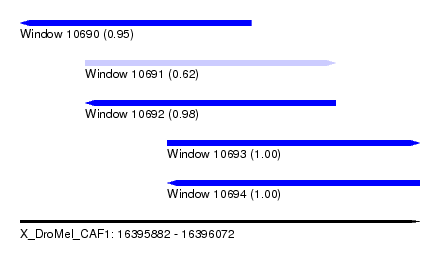
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,395,882 – 16,396,072 |
| Length | 190 |
| Max. P | 0.999914 |

| Location | 16,395,882 – 16,395,992 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -18.36 |
| Energy contribution | -20.18 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16395882 110 - 22224390 UUCAUAGCACGCGUCUAGGCAGUUUUUUAAGUGGUUUCAAAACCCAGAGAAUGAUAUUUAAAGAUAGCCAAGACGUGAUAAAAAAAUACCCAUAAA-AACUAUACUCAAAU .........(((((((.(((...((((((((((.((((........))))....))))))))))..))).)))))))...................-.............. ( -24.50) >DroSec_CAF1 16924 111 - 1 UUCGUAGCACACGUCUAGGCAGUUUUUGAAGUGGUUUCAAAACCCAGAGAAUGAUAUUUAAAGAUAGCCAAGACGUGAUGAAAAAAUGCCCAUAAAAAACUAUACUCAAAU (((((....(((((((.(((.(((((((((....((((........))))......))))))))).))).))))))))))))............................. ( -26.00) >DroSim_CAF1 15307 111 - 1 UUCGUAGCACACGUCUAGGCAGUUUUUGAAGUGGUUUCAAAACCCAGAGAAUGAUAUUUAAAGAUAGCCAAGACGUGAUGAAAAAAUGCCCAUAAAAAACUAUACUCAAAU (((((....(((((((.(((.(((((((((....((((........))))......))))))))).))).))))))))))))............................. ( -26.00) >DroYak_CAF1 13382 96 - 1 --CGUAGCACACAUUUUGGCAGCUUUUGAAGUGU------------GAAAACGAUAUUUAAAGAUAGCCAA-ACGUGAUGAAAAAAUGCCCAUAAGAAACUAUACUCAAAU --.((((..(((..((((((...(((((((.(((------------....)))...)))))))...)))))-).)))(((..........)))......))))........ ( -15.80) >consensus UUCGUAGCACACGUCUAGGCAGUUUUUGAAGUGGUUUCAAAACCCAGAGAAUGAUAUUUAAAGAUAGCCAAGACGUGAUGAAAAAAUGCCCAUAAAAAACUAUACUCAAAU .........(((((((.(((.(((((((((....((((........))))......))))))))).))).))))))).................................. (-18.36 = -20.18 + 1.81)


| Location | 16,395,913 – 16,396,032 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -20.69 |
| Energy contribution | -21.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617028 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16395913 119 + 22224390 AUCACGUCUUGGCUAUCUUUAAAUAUCAUUCUCUGGGUUUUGAAACCACUUAAAAAACUGCCUAGACGCGUGCUAUGAAAGACUUUUCAAAUCAAAAUUGUUUUGUUUUUCAUAUCAUA ....(((((.(((....(((((....((.....))((((....))))..))))).....))).))))).(((.((((((((((....(((.......)))....)))))))))).))). ( -25.70) >DroSec_CAF1 16956 118 + 1 AUCACGUCUUGGCUAUCUUUAAAUAUCAUUCUCUGGGUUUUGAAACCACUUCAAAAACUGCCUAGACGUGUGCUACGAAAGACUUUUCGAAUGAAAAUUU-UUUGUUUUUCAUACCAUA ..(((((((.(((............(((.....))).(((((((.....)))))))...))).)))))))......(((((((.....((((....))))-...)))))))........ ( -26.30) >DroSim_CAF1 15339 118 + 1 AUCACGUCUUGGCUAUCUUUAAAUAUCAUUCUCUGGGUUUUGAAACCACUUCAAAAACUGCCUAGACGUGUGCUACGAAAGACUUUUCGAAUGAAAAUUG-UUUGUUUUUCAUACCAUA ..(((((((.(((............(((.....))).(((((((.....)))))))...))).))))))).....(((((....))))).(((((((...-.....)))))))...... ( -25.40) >consensus AUCACGUCUUGGCUAUCUUUAAAUAUCAUUCUCUGGGUUUUGAAACCACUUCAAAAACUGCCUAGACGUGUGCUACGAAAGACUUUUCGAAUGAAAAUUG_UUUGUUUUUCAUACCAUA ..(((((((.(((.............((.....))((((....))))............))).)))))))......(((((((....(((.......)))....)))))))........ (-20.69 = -21.13 + 0.45)

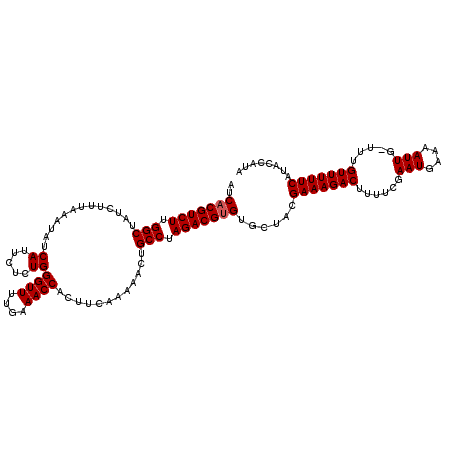
| Location | 16,395,913 – 16,396,032 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 95.51 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.64 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.83 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16395913 119 - 22224390 UAUGAUAUGAAAAACAAAACAAUUUUGAUUUGAAAAGUCUUUCAUAGCACGCGUCUAGGCAGUUUUUUAAGUGGUUUCAAAACCCAGAGAAUGAUAUUUAAAGAUAGCCAAGACGUGAU ..((.(((((((.((....(((.......)))....)).))))))).))(((((((.(((...((((((((((.((((........))))....))))))))))..))).))))))).. ( -30.90) >DroSec_CAF1 16956 118 - 1 UAUGGUAUGAAAAACAAA-AAAUUUUCAUUCGAAAAGUCUUUCGUAGCACACGUCUAGGCAGUUUUUGAAGUGGUUUCAAAACCCAGAGAAUGAUAUUUAAAGAUAGCCAAGACGUGAU ..((.(((((((.((...-...(((((....))))))).))))))).))(((((((.(((.(((((((((....((((........))))......))))))))).))).))))))).. ( -33.60) >DroSim_CAF1 15339 118 - 1 UAUGGUAUGAAAAACAAA-CAAUUUUCAUUCGAAAAGUCUUUCGUAGCACACGUCUAGGCAGUUUUUGAAGUGGUUUCAAAACCCAGAGAAUGAUAUUUAAAGAUAGCCAAGACGUGAU ..((.(((((((.((...-...(((((....))))))).))))))).))(((((((.(((.(((((((((....((((........))))......))))))))).))).))))))).. ( -33.60) >consensus UAUGGUAUGAAAAACAAA_CAAUUUUCAUUCGAAAAGUCUUUCGUAGCACACGUCUAGGCAGUUUUUGAAGUGGUUUCAAAACCCAGAGAAUGAUAUUUAAAGAUAGCCAAGACGUGAU ..((.(((((((.((.......(((((....))))))).))))))).))(((((((.(((.(((((((((....((((........))))......))))))))).))).))))))).. (-30.42 = -30.64 + 0.23)


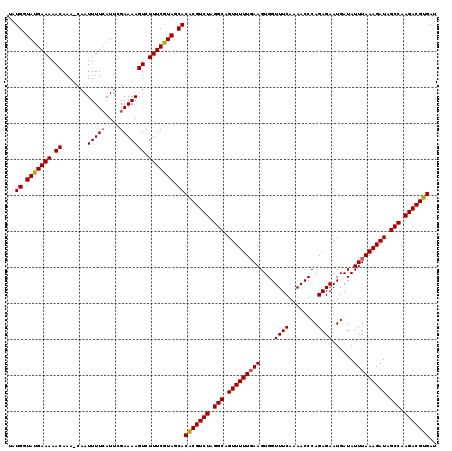
| Location | 16,395,952 – 16,396,072 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -21.44 |
| Energy contribution | -26.00 |
| Covariance contribution | 4.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -4.24 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.52 |
| SVM RNA-class probability | 0.999914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16395952 120 + 22224390 UUGAAACCACUUAAAAAACUGCCUAGACGCGUGCUAUGAAAGACUUUUCAAAUCAAAAUUGUUUUGUUUUUCAUAUCAUACUAAGAGGCAGCUUUCGAAAUGGUUUCAAAACUCAAGGGA (((((((((.((..(((.((((((....(..((.((((((((((....(((.......)))....)))))))))).))..)....)))))).)))..)).)))))))))........... ( -35.60) >DroSec_CAF1 16995 118 + 1 UUGAAACCACUUCAAAAACUGCCUAGACGUGUGCUACGAAAGACUUUUCGAAUGAAAAUUU-UUUGUUUUUCAUACCAUACUGAUAGGCAGCUUUC-AAAUGGUUUCAAAACUCAAGGGA (((((((((.((..(((.(((((((...(((((.((.(((((((.....((((....))))-...))))))).)).)))))...))))))).))).-.)))))))))))........... ( -36.20) >DroSim_CAF1 15378 119 + 1 UUGAAACCACUUCAAAAACUGCCUAGACGUGUGCUACGAAAGACUUUUCGAAUGAAAAUUG-UUUGUUUUUCAUACCAUACUGAUAGGCAGCUUUCGAAAUGGUUUCAAAACUCAAGGGA (((((((((.(((.(((.(((((((...(((((.((.(((((((....(.(((....))))-...))))))).)).)))))...))))))).))).))).)))))))))........... ( -39.40) >DroYak_CAF1 13446 90 + 1 ------ACACUUCAAAAGCUGCCAAAAUGUGUGCUACG------------------------UUUCUUUUUCAUACCAUAUAUAUGGACAGCUUUCGAAAUGGUUUUUAAACUCAAGCGA ------.((.(((.((((((((((..(((((((.((.(------------------------.........).)).))))))).))).))))))).))).))((((........)))).. ( -19.40) >consensus UUGAAACCACUUCAAAAACUGCCUAGACGUGUGCUACGAAAGACUUUUCGAAUGAAAAUUG_UUUGUUUUUCAUACCAUACUGAUAGGCAGCUUUCGAAAUGGUUUCAAAACUCAAGGGA (((((((((.(((.(((.(((((((...(((((.((.(((((((.....................))))))).)).)))))...))))))).))).))).)))))))))........... (-21.44 = -26.00 + 4.56)



| Location | 16,395,952 – 16,396,072 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -39.11 |
| Consensus MFE | -34.03 |
| Energy contribution | -37.28 |
| Covariance contribution | 3.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -5.71 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16395952 120 - 22224390 UCCCUUGAGUUUUGAAACCAUUUCGAAAGCUGCCUCUUAGUAUGAUAUGAAAAACAAAACAAUUUUGAUUUGAAAAGUCUUUCAUAGCACGCGUCUAGGCAGUUUUUUAAGUGGUUUCAA ...........((((((((((((.(((((((((((....(..((.(((((((.((....(((.......)))....)).))))))).))..)....))))))))))).)))))))))))) ( -40.40) >DroSec_CAF1 16995 118 - 1 UCCCUUGAGUUUUGAAACCAUUU-GAAAGCUGCCUAUCAGUAUGGUAUGAAAAACAAA-AAAUUUUCAUUCGAAAAGUCUUUCGUAGCACACGUCUAGGCAGUUUUUGAAGUGGUUUCAA ...........((((((((((((-((((((((((((...((.((.(((((((.((...-...(((((....))))))).))))))).)).))...)))))))))))).)))))))))))) ( -42.00) >DroSim_CAF1 15378 119 - 1 UCCCUUGAGUUUUGAAACCAUUUCGAAAGCUGCCUAUCAGUAUGGUAUGAAAAACAAA-CAAUUUUCAUUCGAAAAGUCUUUCGUAGCACACGUCUAGGCAGUUUUUGAAGUGGUUUCAA ...........(((((((((((((((((((((((((...((.((.(((((((.((...-...(((((....))))))).))))))).)).))...))))))))))))))))))))))))) ( -48.60) >DroYak_CAF1 13446 90 - 1 UCGCUUGAGUUUAAAAACCAUUUCGAAAGCUGUCCAUAUAUAUGGUAUGAAAAAGAAA------------------------CGUAGCACACAUUUUGGCAGCUUUUGAAGUGU------ ..................((((((((((((((.(((......((.((((.........------------------------)))).)).......))))))))))))))))).------ ( -25.42) >consensus UCCCUUGAGUUUUGAAACCAUUUCGAAAGCUGCCUAUCAGUAUGGUAUGAAAAACAAA_CAAUUUUCAUUCGAAAAGUCUUUCGUAGCACACGUCUAGGCAGUUUUUGAAGUGGUUUCAA ............((((((((((((((((((((((((...((.((.(((((((.((.....................)).))))))).)).))...)))))))))))))))))))))))). (-34.03 = -37.28 + 3.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:35 2006