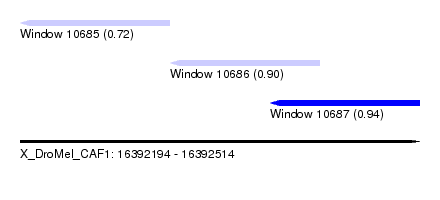
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,392,194 – 16,392,514 |
| Length | 320 |
| Max. P | 0.939109 |

| Location | 16,392,194 – 16,392,314 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -36.54 |
| Energy contribution | -36.23 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16392194 120 - 22224390 UCCGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAAAGCCAGACCACCGGCUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGUGUACAACAACA ...((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...))).(((....)))..))))))))))......................... ( -38.50) >DroVir_CAF1 18857 120 - 1 UACGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAGAGCCAGACCACCGGUUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGUGUACAACAACA ...((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...))).(((....)))..))))))))))......................... ( -38.10) >DroGri_CAF1 10695 120 - 1 UACGGGCGCACGAGGCACAGGACGCCGGCUACCGCAUGUACCGUAAGAGCCAGACCACCGGUUUUCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAAUUAUCUGGACGUGUACAACAACA ...((((((.((((((...(((.(((((((...((.......))...)))).(.....))))..)))...)).(((....)))..))))))))))......................... ( -32.40) >DroSec_CAF1 13280 120 - 1 UCCGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAGAGCCAGACCACCGGCUUCCCCUCGCUGAUCACCAUUUUCUCGGCGCCCAACUAUCUCGACGUGUACAACAACA ...((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...)))((.....))....))))))))))......................... ( -38.60) >DroMoj_CAF1 11165 120 - 1 UACGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAGAGCCAGACCACCGGUUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGUGUACAACAACA ...((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...))).(((....)))..))))))))))......................... ( -38.10) >DroAna_CAF1 8603 120 - 1 UCCGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGGAAGAGCCAGACCACCGGCUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGUGUACAACAACA ...((((((.(((((((.(((...((((.(((.....)))))))..(((((........)))))..)))))).(((....)))..))))))))))......................... ( -41.30) >consensus UACGGGCGCACGAGGCGCAGGACGCCGGCUACCGCAUGUACCGCAAGAGCCAGACCACCGGCUUCCCCUCGCUGAUCACCAUCUUCUCGGCGCCCAACUAUCUCGACGUGUACAACAACA ...((((((.(((((((..(((.(((((((...((.......))...)))).(.....)))).)))...))).(((....)))..))))))))))......................... (-36.54 = -36.23 + -0.30)


| Location | 16,392,314 – 16,392,434 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.89 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -32.98 |
| Energy contribution | -32.70 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16392314 120 - 22224390 ACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCCGUGCGCGGAUGCUCGUACUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCUGCUGUCGAUCA .....((((.((((.....))))...........((((......))))((((..(((.(((....))))))....)))).)))).((((.....((((.........)))).)))).... ( -32.70) >DroVir_CAF1 18977 120 - 1 ACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCGGUGCGUGGCUGUUCGUAUUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAAUCUGCUGUCGAUCA ...((((((.((((.....))))..........((((((....(((((((((((((..........))))))))).)))))).)))).......(((((.......)))))))))))... ( -38.80) >DroGri_CAF1 10815 120 - 1 ACUUCGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCGGUGCGCGGCUGUUCGUAUUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCUGCUGUCGAUCA ...((((((.((((.....))))...........(((......((((((..(((((..........)))))..)).))))..(((((......)))))..........)))))))))... ( -35.50) >DroYak_CAF1 9721 120 - 1 ACUUCGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCCGUGCGCGGCUGCUCGUACUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCUGCUGUCGAUCA ....(((((.((((.....))))...........((((......))))(((((.(((.(((....))))))...))))).)))))((((.....((((.........)))).)))).... ( -36.80) >DroMoj_CAF1 11285 120 - 1 ACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCGGUGCGCGGCUGUUCGUAUUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAAUCUGCUGUCGAUCA ...((((((.((((.....))))..........((((((....((((((..(((((..........)))))..)).)))))).)))).......(((((.......)))))))))))... ( -35.10) >DroAna_CAF1 8723 120 - 1 ACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCGGUGCGCGGGUGCUCGUACUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCUGCUGUCGAUCA ...((((((..(((.....)))(((..((.....((((......))))(((((((..((((((.....)).)))).)))))))...))..))).((((.........))))))))))... ( -34.70) >consensus ACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAACUCGGUGCGCGGCUGCUCGUACUUCUACAGCUACGCCGCCUGCUGCGACUUCCUGCAGAACAACAACCUGCUGUCGAUCA ....(((((.((((.....))))...........((((......))))(((((.(((.(((....))))))...))))).)))))((((.....((((.........)))).)))).... (-32.98 = -32.70 + -0.28)


| Location | 16,392,394 – 16,392,514 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -46.23 |
| Consensus MFE | -42.54 |
| Energy contribution | -42.82 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939109 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16392394 120 - 22224390 CAUCCGGCGGCUCGACCGCUUCAAGGAGCCGCCCGCCUUUGGCCCCAUGUGCGACCUGCUGUGGUCCGAUCCCCUGGAGGACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAA .....((((((((............)))))))).(((...)))....((((((...(((((.(((((..(((...)))))))).))))).((((.....))))..........)))))). ( -45.10) >DroVir_CAF1 19057 120 - 1 CAUAAGGCGGCUCGAUAGAUUCAAGGAGCCGCCCGCCUUUGGCCCCAUGUGCGAUCUGCUGUGGUCCGAUCCGCUCGAGGACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAA .....((((((((............)))))))).(((...)))....((((((...((..(.((((((((((......))).((((....))))......))))))).)..)))))))). ( -44.30) >DroGri_CAF1 10895 120 - 1 CAUUAGGCGGCUCGAUAGAUUCAAGGAGCCGCCCGCCUUUGGCCCCAUGUGCGACUUGCUGUGGUCCGAUCCGCUCGAGGACUUCGGCAACGAGAAGAACUCGGACUUCUACACGCACAA .....((((((((............)))))))).(((...)))....((((((..((((((.(((((...........))))).))))))((((.....))))..........)))))). ( -46.80) >DroSec_CAF1 13480 120 - 1 CAUACGGCGGCUCGACCGCUUCAAGGAGCCGCCCGCCUUCGGCCCCAUGUGCGACCUGCUGUGGUCCGAUCCGCUGGAGGACUUCGGCAACGAGAAGAACUCGGACUUCUACACGCACAA .....((((((((............)))))))).(((...)))....((((((...(((((.(((((..(((...)))))))).))))).((((.....))))..........)))))). ( -47.40) >DroMoj_CAF1 11365 120 - 1 CAUUAGGCGGCUGGAUAGAUUCAAGGAGCCGCCCGCCUUUGGCCCCAUGUGCGACCUGCUGUGGUCCGAUCCGCUCGAGGACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAA .....(((((((.((.....))....))))))).(((...)))....((((((...((..(.((((((((((......))).((((....))))......))))))).)..)))))))). ( -42.00) >DroAna_CAF1 8803 120 - 1 UAUCAGGCGGCUGGAUAGGUUCAAGGAGCCGCCGGCCUUCGGGCCCAUGUGCGACCUGCUGUGGUCCGAUCCGCUGGAGGACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAA .....(((((((.((.....))....)))))))((((....))))..((((((...(((((.(((((..(((...)))))))).))))).((((.....))))..........)))))). ( -51.80) >consensus CAUAAGGCGGCUCGAUAGAUUCAAGGAGCCGCCCGCCUUUGGCCCCAUGUGCGACCUGCUGUGGUCCGAUCCGCUCGAGGACUUUGGCAACGAGAAGAACUCGGACUUCUACACGCACAA .....((((((((............)))))))).(((...)))....((((((...((..(.((((((((((......))).((((....))))......))))))).)..)))))))). (-42.54 = -42.82 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:29 2006