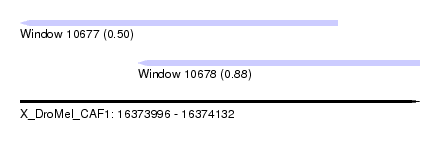
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,373,996 – 16,374,132 |
| Length | 136 |
| Max. P | 0.878251 |

| Location | 16,373,996 – 16,374,104 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -41.05 |
| Consensus MFE | -25.38 |
| Energy contribution | -25.33 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16373996 108 - 22224390 UCUUUGGCCAG--------ACAGUUG----GCCACUCCAGCUGGCCCUGUUCUAUACGGCACUUUCGCCCAGUGGCGGACUGGCCAAUGUGUGCGCCGUCUUCCUUAAGGGAAAUAUCAA .....(((((.--------(((.(((----((((.(((.((((.............))))......(((....)))))).)))))))))).)).)))...((((.....))))....... ( -40.02) >DroSec_CAF1 719 108 - 1 UCUUUGGCCAG--------GCAGUUG----GCCACUCCAGCUGGCCCUCUUCUAUACGGCACUGGCGCCCAGUGGCGGACUGGCCAAUGUGUGCGCCGUCUUCCUCAAGGGAAAUAUCAA .....(((((.--------(((.(((----((((.(((.(((.(((...........)))...)))(((....)))))).)))))))))).)).)))...((((.....))))....... ( -42.00) >DroEre_CAF1 711 108 - 1 UCUUUGGCCAG--------CUAGUUG----GCCACUCCAGCUGGCCCUCUUCUACACGGCACUGGCGCCCAGUGGCGGCCUGGCCAAUGUGUGCGCCGUCUUCCUCAAGGGAAAUAUCAA ...((((((((--------(((((((----(.....)))))))))...........((.((((((...)))))).))...))))))).............((((.....))))....... ( -39.80) >DroWil_CAF1 714 120 - 1 CUUGUGGUCAUCGUCCUCCGCAAAUGAGCAGUCCCUGCAAUUGGCAUUGUUCUACACGGCUCUGGCACCCAGUGGCGGGUUGGCCAAUGUCUGUGCCGUUUUCCUAAAGGGCAAUGUGAA .((((((..........))))))..(((((((.((.......)).)))))))..((((((..((((((((......))))..))))..)))..((((.(((....))).))))..))).. ( -39.90) >DroYak_CAF1 830 108 - 1 UCUGUGGCCAG--------GUAGUUG----GCCACUGCAGCUGGCCCUCUUCUACACGGCACUGGCGCCCAGUGGCGGACUGGCCAAUGUGUGCGCCGUCUUUCUCAAGGGAAAUAUCAA ....(((((((--------((((..(----(((((....).))))).....)))).((.((((((...)))))).))..)))))))..............(((((....)))))...... ( -39.90) >DroAna_CAF1 26572 108 - 1 CCUCUGGCCGG--------GGCAAUG----GCCCCUCCAGCUGGCCCUGUUCUACACGGCCUUGGCGCCCAGCGGCGGAUUGGCCAAUGUCUGCACCGUUUUCUGUAAAGGAAACCUCAA ....(((((((--------(((....----)))))(((.(((((((.((.....)).))))..)))(((....))))))..)))))...........(((((((....)))))))..... ( -44.70) >consensus UCUUUGGCCAG________GCAGUUG____GCCACUCCAGCUGGCCCUCUUCUACACGGCACUGGCGCCCAGUGGCGGACUGGCCAAUGUGUGCGCCGUCUUCCUCAAGGGAAAUAUCAA ....(((((((...................((((.......))))...........((.(((((.....))))).))..)))))))...........((.((((....)))).))..... (-25.38 = -25.33 + -0.05)
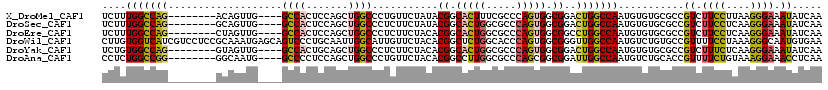
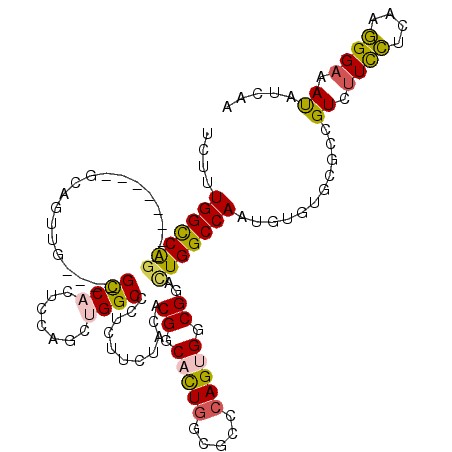
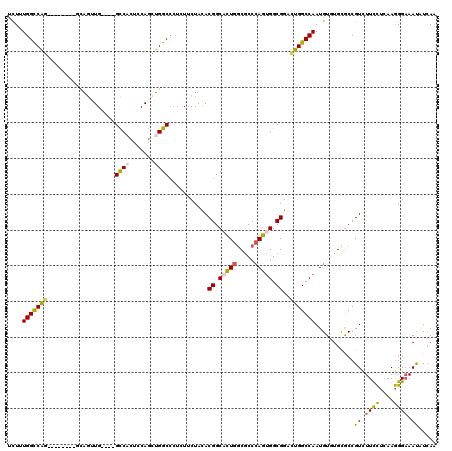
| Location | 16,374,036 – 16,374,132 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -46.13 |
| Consensus MFE | -26.00 |
| Energy contribution | -26.51 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16374036 96 - 22224390 CCGCCGUGGGAUUCGGACUGGGUCGUGCUCUUUGGCCAG--------ACAGUUG----GCCACUCCAGCUGGCCCUGUUCUAUACGGCACUUUCGCCCAGUGGCGGAC ((((((((((....((((.((((((.(((...(((((((--------....)))----))))....))))))))).))))....(((.....))))))).)))))).. ( -44.60) >DroSec_CAF1 759 96 - 1 CCGCCUUGGGAUUCGGACUGGGUCGUGCUCUUUGGCCAG--------GCAGUUG----GCCACUCCAGCUGGCCCUCUUCUAUACGGCACUGGCGCCCAGUGGCGGAC ((((((((((....(((..((((((.(((...(((((((--------....)))----))))....)))))))))..)))......((....)).))))).))))).. ( -43.30) >DroEre_CAF1 751 96 - 1 CCGCCUUGGCCUUUGGACUGGGUCGUGCUCUUUGGCCAG--------CUAGUUG----GCCACUCCAGCUGGCCCUCUUCUACACGGCACUGGCGCCCAGUGGCGGCC (((((...(((..((((..((((((.(((...(((((((--------....)))----))))....)))))))))...))))...)))(((((...)))))))))).. ( -44.90) >DroWil_CAF1 754 108 - 1 CAGCCCUGGCCUUUGGCCUGGGACGAGCCUUGUGGUCAUCGUCCUCCGCAAAUGAGCAGUCCCUGCAAUUGGCAUUGUUCUACACGGCUCUGGCACCCAGUGGCGGGU ..(((..((((....((..((((((((((....)))..)))))))..))....(((((((.((.......)).))))))).....))))..)))((((......)))) ( -42.70) >DroYak_CAF1 870 96 - 1 CCGCCCUGGGAUUCGGACUGGGUCGUGCUCUGUGGCCAG--------GUAGUUG----GCCACUGCAGCUGGCCCUCUUCUACACGGCACUGGCGCCCAGUGGCGGAC ((((((((((....(((..((((((.((((.((((((((--------....)))----))))).).)))))))))..)))......((....)).))))).))))).. ( -50.50) >DroAna_CAF1 26612 96 - 1 CCGGUCUGGGCUUUGGUCUGGGUCGUGCCCUCUGGCCGG--------GGCAAUG----GCCCCUCCAGCUGGCCCUGUUCUACACGGCCUUGGCGCCCAGCGGCGGAU (((.(((((((..(((...((((((((((((......))--------))).)))----))))..)))(((((((.((.....)).))))..))))))))).).))).. ( -50.80) >consensus CCGCCCUGGGAUUCGGACUGGGUCGUGCUCUUUGGCCAG________GCAGUUG____GCCACUCCAGCUGGCCCUCUUCUACACGGCACUGGCGCCCAGUGGCGGAC ((((((((((...((..((((((((((......((((((.............................))))))........)))))).))))))))))).))))).. (-26.00 = -26.51 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:20 2006