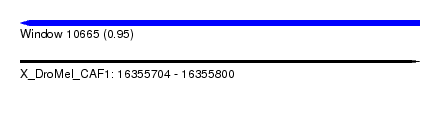
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,355,704 – 16,355,800 |
| Length | 96 |
| Max. P | 0.951834 |

| Location | 16,355,704 – 16,355,800 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.05 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16355704 96 - 22224390 UUGCAUACUGUAUGAAACAUGAG---UAA--------CAUCUUCAUUUUCUUCUCUUGCAGGCCUUGGAAAUCUAAGGCUCUCAUCGUUGGCCGCAUAGACAAUGAC .((((....(...((((.(((((---...--------....)))))))))..)...))))((((((((....))))))))....((((((..(.....).)))))). ( -22.90) >DroPse_CAF1 18972 96 - 1 UUACUUACU--------AAUGAC---CGACAUCGAUUUAUGUUUCUUUCGAUUUGUUGCAGGCCUUGGAAAUCUAAGGCUCUCAUCGUUGGCCGCAUAGACAAUGAC .......((--------(.((.(---(((((((((............)))))..(.((..((((((((....))))))))..)).))))))...)))))........ ( -23.70) >DroSec_CAF1 22954 99 - 1 UUGCGUACUGUACUGUACAUGAGACAUAA--------UAUCUUCAUUUUGCUCUCUUGCAGGCCUUGGAAAUCUAAGGCUCUCAUCGUUGGCCGCAUAGACAAUGAC ....((((......)))).(((((.....--------..........((((......))))(((((((....))))))))))))((((((..(.....).)))))). ( -27.40) >DroSim_CAF1 17407 99 - 1 UUGCGUACUGUACUGUACACGAGACAUAA--------UAUCUUCCUUUUGUUCUCUUGCAGGCCUUGGAAAUCUAAGGCUCUCAUCGUUGGCCGCAUAGACAAUGAC .(((((((......))))..((((.((((--------..........))))))))..)))((((((((....))))))))....((((((..(.....).)))))). ( -26.60) >DroEre_CAF1 17060 96 - 1 CUGCAUAUUGUCUGAAACAUGAC---UAA--------CAGCUACCCUCUGUUCUUUUGCAGGCCUUGGAAAUCUAAGGCUCUCAUCGUUGGCCGCAUAGACAAUGAC .....(((((((((.....((.(---(((--------(.........((((......))))(((((((....))))))).......))))).))..))))))))).. ( -26.90) >DroYak_CAF1 16369 77 - 1 -------------------UUAG---UAA--------UAUUUUCCCUCUAUUCUUUCACAGGCCUUGGAAAUCUAAGGCUCUCAUCGUUGGCCGCAUAGACAAUGAC -------------------....---...--------.......................((((((((....))))))))....((((((..(.....).)))))). ( -15.80) >consensus UUGCAUACUGUA____ACAUGAG___UAA________UAUCUUCCUUUUGUUCUCUUGCAGGCCUUGGAAAUCUAAGGCUCUCAUCGUUGGCCGCAUAGACAAUGAC ............................................................((((((((....))))))))....((((((..(.....).)))))). (-15.80 = -15.80 + -0.00)


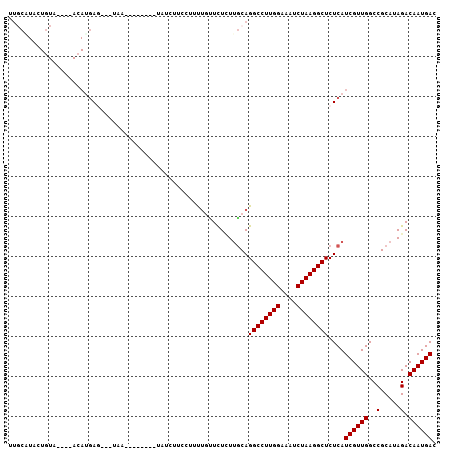
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:08 2006