
| Sequence ID | X_DroMel_CAF1 |
|---|---|
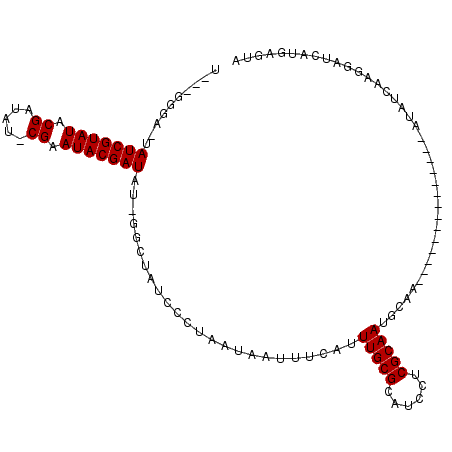
| Location | 16,353,201 – 16,353,299 |
| Length | 98 |
| Max. P | 0.834814 |

| Location | 16,353,201 – 16,353,299 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16353201 98 + 22224390 UACGGGAAUAAUCGUAUACGAUAU-CGAAUACGAUAUUGGCUAUCCCUAAUAAUUUCAUUUGCGCAUCCUCGCAAUGCAA--------------AUAUCAUGAAUGAUGAGCA (((((......)))))..((((((-((....))))))))(((.................(((((......))))).....--------------.(((((....)))))))). ( -20.60) >DroSec_CAF1 20547 94 + 1 U---GGGA-UAUCGUAUACGAUAU-CGGAUACGAUAUCGGCUAUCCCUAAUAACUUCAUUUGCGCAUCCUCGCAAUGCAA--------------AUAUCAAGGAUCAUGAGUA .---((((-((..((...((((((-((....))))))))))))))))......(((.((((((((......))...))))--------------))...)))........... ( -23.40) >DroYak_CAF1 13631 100 + 1 U---GGGA-UAUCGUAUACGAUAUUCGAAUACGAUAU---------CUAACAAUUUCAUUUGCGCAUCCUCGCAAUGCAAAAUGCAAAAUGCAAAAUUCAAGGAUCAUGUUUA .---.(((-((((((((.((.....)).)))))))))---------))..........(((((((......))..(((.....)))....))))).................. ( -24.60) >consensus U___GGGA_UAUCGUAUACGAUAU_CGAAUACGAUAU_GGCUAUCCCUAAUAAUUUCAUUUGCGCAUCCUCGCAAUGCAA______________AUAUCAAGGAUCAUGAGUA ..........(((((((.((.....)).)))))))........................(((((......)))))...................................... (-12.00 = -12.00 + 0.00)


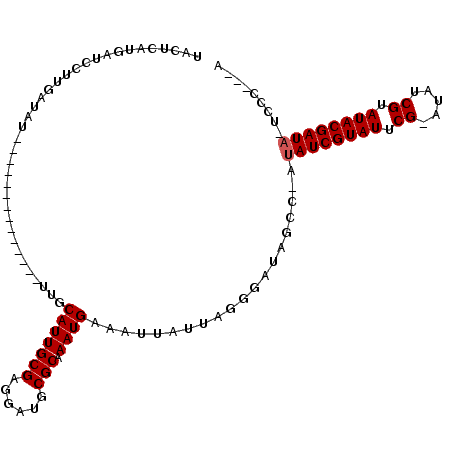
| Location | 16,353,201 – 16,353,299 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 74.06 |
| Mean single sequence MFE | -27.27 |
| Consensus MFE | -12.53 |
| Energy contribution | -12.87 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16353201 98 - 22224390 UGCUCAUCAUUCAUGAUAU--------------UUGCAUUGCGAGGAUGCGCAAAUGAAAUUAUUAGGGAUAGCCAAUAUCGUAUUCG-AUAUCGUAUACGAUUAUUCCCGUA (((.((((.(((..(((..--------------....)))..))))))).))).............((((........(((((((.((-....)).)))))))...))))... ( -23.30) >DroSec_CAF1 20547 94 - 1 UACUCAUGAUCCUUGAUAU--------------UUGCAUUGCGAGGAUGCGCAAAUGAAGUUAUUAGGGAUAGCCGAUAUCGUAUCCG-AUAUCGUAUACGAUA-UCCC---A ........((((((((((.--------------...(((((((......))).))))....))))))))))....((((((((((.((-....)).))))))))-))..---. ( -31.90) >DroYak_CAF1 13631 100 - 1 UAAACAUGAUCCUUGAAUUUUGCAUUUUGCAUUUUGCAUUGCGAGGAUGCGCAAAUGAAAUUGUUAG---------AUAUCGUAUUCGAAUAUCGUAUACGAUA-UCCC---A ........(((((((.....((((..........))))...)))))))..((((......))))..(---------(((((((((.((.....)).))))))))-))..---. ( -26.60) >consensus UACUCAUGAUCCUUGAUAU______________UUGCAUUGCGAGGAUGCGCAAAUGAAAUUAUUAGGGAUAGCC_AUAUCGUAUUCG_AUAUCGUAUACGAUA_UCCC___A ....................................(((((((......))).))))....................((((((((.((.....)).))))))))......... (-12.53 = -12.87 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:06 2006