
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,351,576 – 16,351,734 |
| Length | 158 |
| Max. P | 0.999889 |

| Location | 16,351,576 – 16,351,681 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.58 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -12.31 |
| Energy contribution | -13.47 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.35 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16351576 105 + 22224390 UUCUUAGCCACUCGAGGCACUCAGUCAUUUCGGG--------------GACUCCACCUUGAUAAGUAAUU-GAAGCCCCCGGGCAGUGUCAAUAACUAUGGCAAUGCCAAAUGGCAAGCG ......(((......))).....(((((((((((--------------(.((.((..............)-).)).)))))((((.(((((.......))))).)))))))))))..... ( -32.04) >DroPse_CAF1 14456 115 + 1 GCCUUAGCCA----GGGCACUCAGUCAUAUUGAGUGUACUCCCAGCUGGGAGCCACCUUGAUAAGUAAUU-AAAGCCCGCAGGCAAUGCCAAUAGCUAUGGCAAUGACAAAUGCCAAAUA (((.((((((----(((((((((((...))))))))..(((((....)))))...)))))..........-...(((....)))..........)))).))).................. ( -36.00) >DroSec_CAF1 18975 105 + 1 UCCUUAGCCACUCGAGGCACUCAGUCAUUUCGGG--------------GGCUCCACCUUGAUAAGUAAUU-GAAGCCCCCGAGCAGUGUCAAUAACUAUGGCAAUGCCAAAUGGCAAGCG ......(((......))).....(((((((((((--------------((((.((..............)-).)))))))))(((.(((((.......))))).)))..))))))..... ( -35.74) >DroEre_CAF1 13092 90 + 1 UCCUUAGCCA---------------CAUUUCGGG--------------GGCUCCACCUCGAUAAGUAAUU-GAAGCCCCCAGGCAGUGUCAAUAACUAUGACAAUGCCAAAUGGCAAGCG ..(((.((((---------------......(((--------------((((.....(((((.....)))-))))))))).((((.(((((.......))))).))))...))))))).. ( -35.50) >DroPer_CAF1 14636 116 + 1 GCCUUAGCCA----GGGCACUCAGUCAUAUUGAGUGUACUCCCAGCUGGGAGCCACCUUGAUAAGUAAAUAAAAGCCCGCAGGCAAUGCCAAUAACUAUGGCAAUGANNNNNNNNNNNNN ((((..((..----(((((((((((...)))))))...(((((....)))))......................))))))))))..(((((.......)))))................. ( -34.30) >consensus UCCUUAGCCA____AGGCACUCAGUCAUUUCGGG______________GGCUCCACCUUGAUAAGUAAUU_GAAGCCCCCAGGCAGUGUCAAUAACUAUGGCAAUGCCAAAUGGCAAGCG ..(((((((......)))...........(((((......................))))))))).........(((....((((.(((((.......))))).))))....)))..... (-12.31 = -13.47 + 1.16)


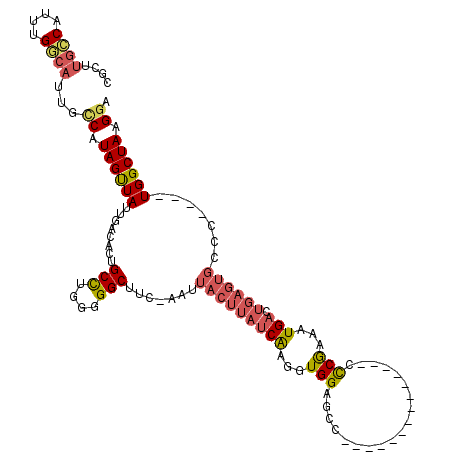
| Location | 16,351,576 – 16,351,681 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.58 |
| Mean single sequence MFE | -40.46 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.55 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.34 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16351576 105 - 22224390 CGCUUGCCAUUUGGCAUUGCCAUAGUUAUUGACACUGCCCGGGGGCUUC-AAUUACUUAUCAAGGUGGAGUC--------------CCCGAAAUGACUGAGUGCCUCGAGUGGCUAAGAA ..((((((((((((((((....((((((((.........((((((((((-(....((.....)).)))))))--------------)))).))))))))))))))..))))))).))).. ( -40.40) >DroPse_CAF1 14456 115 - 1 UAUUUGGCAUUUGUCAUUGCCAUAGCUAUUGGCAUUGCCUGCGGGCUUU-AAUUACUUAUCAAGGUGGCUCCCAGCUGGGAGUACACUCAAUAUGACUGAGUGCCC----UGGCUAAGGC ....((((....))))..(((.((((((..(((...(((....)))...-....((((((((.((((((((((....)))))).)))).....))).)))))))).----)))))).))) ( -40.30) >DroSec_CAF1 18975 105 - 1 CGCUUGCCAUUUGGCAUUGCCAUAGUUAUUGACACUGCUCGGGGGCUUC-AAUUACUUAUCAAGGUGGAGCC--------------CCCGAAAUGACUGAGUGCCUCGAGUGGCUAAGGA ..((((((((((((((((....((((((((........(((((((((((-(....((.....)).)))))))--------------)))))))))))))))))))..))))))).))).. ( -45.10) >DroEre_CAF1 13092 90 - 1 CGCUUGCCAUUUGGCAUUGUCAUAGUUAUUGACACUGCCUGGGGGCUUC-AAUUACUUAUCGAGGUGGAGCC--------------CCCGAAAUG---------------UGGCUAAGGA ..((((((((..((((.(((((.......))))).))))((((((((((-(....(((...))).)))))))--------------))))....)---------------)))).))).. ( -41.30) >DroPer_CAF1 14636 116 - 1 NNNNNNNNNNNNNUCAUUGCCAUAGUUAUUGGCAUUGCCUGCGGGCUUUUAUUUACUUAUCAAGGUGGCUCCCAGCUGGGAGUACACUCAAUAUGACUGAGUGCCC----UGGCUAAGGC ..................(((.((((((..(((...(((....)))........((((((((.((((((((((....)))))).)))).....))).)))))))).----)))))).))) ( -35.20) >consensus CGCUUGCCAUUUGGCAUUGCCAUAGUUAUUGACACUGCCUGGGGGCUUC_AAUUACUUAUCAAGGUGGAGCC______________CCCGAAAUGACUGAGUGCCC____UGGCUAAGGA ....((((....))))...((.((((((........(((....))).......(((((((((...(((...................)))...))).)))))).......)))))).)). (-13.62 = -14.55 + 0.93)


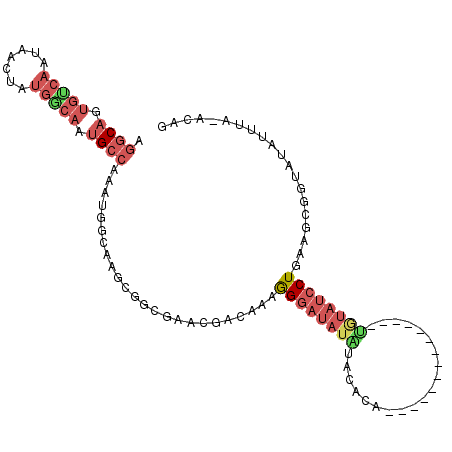
| Location | 16,351,641 – 16,351,734 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 64.96 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -9.21 |
| Energy contribution | -11.21 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.571010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16351641 93 + 22224390 GGGCAGUGUCAAUAACUAUGGCAAUGCCAAAUGGCAAGCGGCGAACGACAAAGGGAUAUAUACACA--------------UGUAUCCUGAAGCGGUAUAUUUA-ACAG .((((.(((((.......))))).))))..((.((..(((.....)).)...(((((((((....)--------------))))))))...)).)).......-.... ( -24.60) >DroPse_CAF1 14531 106 + 1 AGGCAAUGCCAAUAGCUAUGGCAAUGACAAAUGCCAAAUAACAAAGGACAGAGGACGAAGGACAAAGGCUCUUUGUGCCCCAAAAGCUUUGGCUUUAAAUU-G-AUAG .......(((((.((((.(((((........)))))..............(.((((((((..(....)..)))))).)).)...)))))))))........-.-.... ( -26.40) >DroSec_CAF1 19040 94 + 1 GAGCAGUGUCAAUAACUAUGGCAAUGCCAAAUGGCAAGCGGCGAACGACAAAGGGAUAUAUACACA--------------UGUAUCCUGAAGCGGUAUAUUUAUACAG ..((..(((((.......))))).((((....)))).))......((.....(((((((((....)--------------))))))))....))((((....)))).. ( -23.70) >DroEre_CAF1 13142 93 + 1 AGGCAGUGUCAAUAACUAUGACAAUGCCAAAUGGCAAGCGGCGAAAGACAAAGGGAUAUGUACACG--------------UGUAUCCUGAAGCGGUAUAUAUA-ACAG .((((.(((((.......))))).))))..((.((....(.(....).)...((((((..(....)--------------..))))))...)).)).......-.... ( -24.50) >DroAna_CAF1 15485 77 + 1 CGGCUG------------CGGCAAUGCCACACA-CAAG-----GAUCACAAAAGGAUAUUUACGCA------------CUCAUAUCCUUUGGCGUUAUAUUUC-AAAG ..((..------------..))((((((.....-...(-----.....).(((((((((.......------------...))))))))))))))).......-.... ( -15.20) >consensus AGGCAGUGUCAAUAACUAUGGCAAUGCCAAAUGGCAAGCGGCGAACGACAAAGGGAUAUAUACACA______________UGUAUCCUGAAGCGGUAUAUUUA_ACAG .((((.(((((.......))))).))))........................((((((((....................)))))))).................... ( -9.21 = -11.21 + 2.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:03 2006