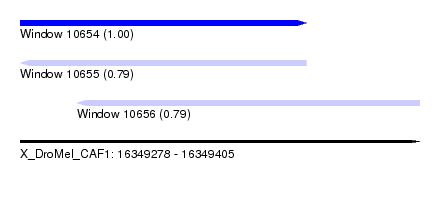
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,349,278 – 16,349,405 |
| Length | 127 |
| Max. P | 0.999648 |

| Location | 16,349,278 – 16,349,369 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -16.62 |
| Consensus MFE | -9.79 |
| Energy contribution | -9.35 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.59 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16349278 91 + 22224390 CAUAUAUAUAUAUAAAACAUAUACUAUGCAUAU--AGCUAUGGCAUAGUUUAGCUAUACCAUAUAGCUGCGACCCACUAUAUGUACACGAACA ......((((((((........((((((((((.--...))).))))))).((((((((...))))))))........))))))))........ ( -18.30) >DroSec_CAF1 16720 85 + 1 CAUAU------AUAAAACAUAUACUAUGCAUAU--AGCUAUGGCAUAGUUUAGCUAUACUAUAUAGCUGCGACCCACUAUAUGUACACGAACA .....------.....((((((((((((((((.--...))).))))))..((((((((...))))))))........)))))))......... ( -17.90) >DroEre_CAF1 10969 72 + 1 CAUAU------AUAAAACAUAUACUAUGCAUAUAC---------------UAGCUAUGCCAUGUAGCUGCGACCCAUUAUAUGUACACGAACA .....------.....(((((((..(((.......---------------(((((((.....))))))).....))))))))))......... ( -11.00) >DroYak_CAF1 12136 80 + 1 CAUAU------AUAUAGUAUAUAUAAAACAUAUAUAGCUAUAGC----UAUAGCUAUAAUAUAUAGCUGCGACCCACUAUAUGUACGCGA--- ..(((------(((((((..............((((((....))----))))((((((...))))))........)))))))))).....--- ( -19.30) >consensus CAUAU______AUAAAACAUAUACUAUGCAUAU__AGCUAUGGC____UUUAGCUAUACCAUAUAGCUGCGACCCACUAUAUGUACACGAACA ................(((((((.............((....))......(((((((.....)))))))........)))))))......... ( -9.79 = -9.35 + -0.44)



| Location | 16,349,278 – 16,349,369 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -9.80 |
| Energy contribution | -9.30 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16349278 91 - 22224390 UGUUCGUGUACAUAUAGUGGGUCGCAGCUAUAUGGUAUAGCUAAACUAUGCCAUAGCU--AUAUGCAUAGUAUAUGUUUUAUAUAUAUAUAUG ......(((.((((((((.(((....))).((((((((((.....)))))))))))))--))))).)))((((((((.....))))))))... ( -25.20) >DroSec_CAF1 16720 85 - 1 UGUUCGUGUACAUAUAGUGGGUCGCAGCUAUAUAGUAUAGCUAAACUAUGCCAUAGCU--AUAUGCAUAGUAUAUGUUUUAU------AUAUG ....((((((((((((((........))))))).(((((((((..........)))))--)))).....)))))))......------..... ( -20.10) >DroEre_CAF1 10969 72 - 1 UGUUCGUGUACAUAUAAUGGGUCGCAGCUACAUGGCAUAGCUA---------------GUAUAUGCAUAGUAUAUGUUUUAU------AUAUG ....((((((((((((....((.(((..(((.((((...))))---------------)))..)))))..))))))).....------))))) ( -12.80) >DroYak_CAF1 12136 80 - 1 ---UCGCGUACAUAUAGUGGGUCGCAGCUAUAUAUUAUAGCUAUA----GCUAUAGCUAUAUAUGUUUUAUAUAUACUAUAU------AUAUG ---...((((.((((((((......((((((((((((((((....----))))))).)))))).))).......))))))))------.)))) ( -20.12) >consensus UGUUCGUGUACAUAUAGUGGGUCGCAGCUAUAUAGUAUAGCUAAA____GCCAUAGCU__AUAUGCAUAGUAUAUGUUUUAU______AUAUG .....(((..(((...)))...)))((((((.....))))))..................((((((...)))))).................. ( -9.80 = -9.30 + -0.50)



| Location | 16,349,296 – 16,349,405 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.38 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -14.16 |
| Energy contribution | -13.85 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16349296 109 - 22224390 CUUCUG----CCACUGGCAUAUACUCGUACUCGUUCUUCAUGUUCGUGUACAUAUAGUGGGUCGCAGCUAUAUGGUAUAGCUAAACUAUGCCAUAGCU--AUAUGCAUAGUAUAU ...(((----(.(((.((.((((...((((.((..(.....)..)).)))).)))))).))).))))...((((((((((.....))))))))))(((--((....))))).... ( -32.20) >DroSec_CAF1 16732 113 - 1 CUUCUGCCUGCCAGUGGCAUAUACUCGUACUCGUUCCCCAUGUUCGUGUACAUAUAGUGGGUCGCAGCUAUAUAGUAUAGCUAAACUAUGCCAUAGCU--AUAUGCAUAGUAUAU ....((((.......))))((((((.((((.((..(.....)..)).))))(((((((........))))))))))))).....(((((((.(((...--.)))))))))).... ( -29.00) >DroEre_CAF1 10981 89 - 1 CCUCUG----C-UCUGGCAUAUACUCG------UUCUCCCUGUUCGUGUACAUAUAAUGGGUCGCAGCUACAUGGCAUAGCUA---------------GUAUAUGCAUAGUAUAU ....((----(-(...(((((((((.(------((..((.(((..(((..(((...)))...)))....))).))...))).)---------------))))))))..))))... ( -21.40) >DroYak_CAF1 12148 98 - 1 CUUCUG----CCUGUGGCAUAUACUCG------UUCUCUA---UCGCGUACAUAUAGUGGGUCGCAGCUAUAUAUUAUAGCUAUA----GCUAUAGCUAUAUAUGUUUUAUAUAU .....(----(.((((((...((((((------(......---..))).......)))).))))))))((((((..(((..((((----((....))))))..)))..)))))). ( -22.31) >consensus CUUCUG____CCACUGGCAUAUACUCG______UUCUCCAUGUUCGUGUACAUAUAGUGGGUCGCAGCUAUAUAGUAUAGCUAAA____GCCAUAGCU__AUAUGCAUAGUAUAU ................((((((.......................(((..(((...)))...)))((((((.....))))))..................))))))......... (-14.16 = -13.85 + -0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:16:00 2006