| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,348,960 – 16,349,054 |
| Length | 94 |
| Max. P | 0.620067 |

| Location | 16,348,960 – 16,349,054 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -9.24 |
| Energy contribution | -10.93 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
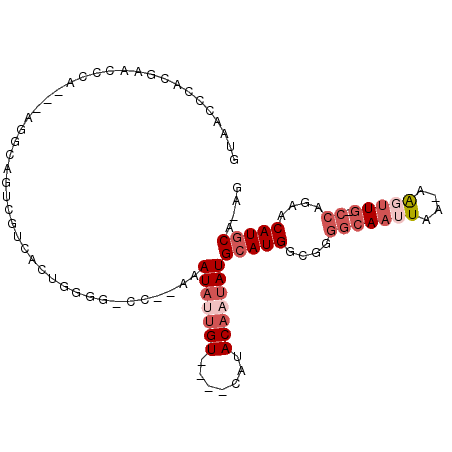
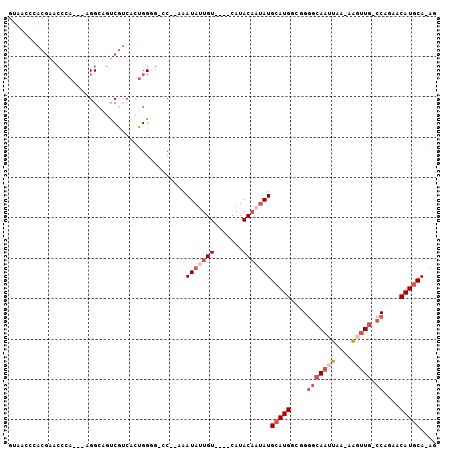
>X_DroMel_CAF1 16348960 94 - 22224390 GUAACCCACGAACCCA---AGGCAGUCGUCACUGGAG-CC--AAAUAUUGU----CAUACAAUAUGCAUGGUGGGGCAAUUAAAAAGUUG-CCAGAACAUGCA-AG ................---.(((..(((....))).)-))--..((((((.----....))))))(((((....(((((((....)))))-))....))))).-.. ( -24.00) >DroSec_CAF1 16404 93 - 1 GUAACCCACGAACCCA---AGGCAGUCGUCACUGGGG-CC--AAAUAUUGU----CAUACAAUAUGCAUGGCGGGGCAAUUAA-AAGUUG-CCAGAACAUGCA-AG (((..((.....((((---.(((....)))..))))(-((--(.((((((.----....))))))...))))))((((((...-..))))-))......))).-.. ( -29.00) >DroSim_CAF1 11599 92 - 1 GUAACCCACGAACCCA---AGGCGGUCGUCACUGGGG-CC--AAAUAUUGU----CAUACA-UAUGCAUGGCGGGGCAAUUAA-AAGUUG-CCAGAACAUGCA-AG .........(..((((---.(((....)))..)))).-.)--.........----......-..((((((....((((((...-..))))-))....))))))-.. ( -26.30) >DroEre_CAF1 10632 93 - 1 GUAACCCACGAGCCCA---AGGCAGUCGUCACUGGGG-CC--AAAUAUUGU----CAUACUAUAUGCAUGGCGGGGCAAUUAA-AAGUUU-GCAGAACAUGCA-AG (((.(((..(.((((.---.(((....)))....)))-))--.......((----(((.(.....).))))))))((((....-....))-))......))).-.. ( -24.30) >DroMoj_CAF1 11876 85 - 1 -----------G-CCA---UGGCC-CCAGCAAUGGGACAC--AAAUAUUGUCCCCCAUACAAUAUGCAUGACAGGGCAAUUAA-AGU-UGCCCAAAGCAUACA-AA -----------(-(((---((..(-(((....))))....--..(((((((.......))))))).))))...(((((((...-.))-)))))...)).....-.. ( -26.80) >DroAna_CAF1 11437 85 - 1 ---------GAACCGAAACAGGAAGCC------GGAG-GCAAAAAUAAUGU----CAUACAAAAUGCAUGGCGGUGCAAUUAA-AAUUUGCCCAGAACAUGCACAA ---------...((......))..(((------...)-))...........----.........((((((..((.((((....-...))))))....))))))... ( -18.20) >consensus GUAACCCACGAACCCA___AGGCAGUCGUCACUGGGG_CC__AAAUAUUGU____CAUACAAUAUGCAUGGCGGGGCAAUUAA_AAGUUG_CCAGAACAUGCA_AG ............................................(((((((.......)))))))(((((....(((((((....))))).))....))))).... ( -9.24 = -10.93 + 1.69)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:58 2006