| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,339,156 – 16,339,269 |
| Length | 113 |
| Max. P | 0.995666 |

| Location | 16,339,156 – 16,339,269 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -17.09 |
| Energy contribution | -17.03 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16339156 113 - 22224390 -ACU--AUCACAGUUGUGGUAGAUAAUU-UCAAUUUCGAUUGUUGCCGGAACU-CGAUUGAAUAUUGAGAAUAUCUACCCGUACCAAUGACUCGAAUACAGCAGCAAC-CACAACAGAC -...--......(((((((((((((.((-(((((((((((((...........-)))))))).))))))).)))))...........((.(((.......).))))))-)))))).... ( -27.90) >DroPse_CAF1 1217 101 - 1 AACUCAAGAACAGCCCUGGUAGAUAAUU-UCAAUUUCGAUUGCUGGGCGAACA-CAAUCGAUUAUUGAGAAUACCUAUC----------ACCUGACU-----AUCAAC-CACAACAGAC ..........(((...((((((.((.((-(((((.(((((((.((......))-)))))))..))))))).)).)))))----------).)))...-----......-.......... ( -23.80) >DroEre_CAF1 1097 113 - 1 -ACU--ACAACAGCUGUGGUAGAUAAUU-UCAAUUUCGAUUGCUGCCGGAACU-CGAUCGAAUAUUGAGAAUAUCUACCCGUACCAAUGACUCGAUAACAGCAGCAAC-CACAACAGAC -...--......(((((((((((((.((-(((((((((((((..(......).-)))))))).))))))).))))))))(((....)))...........)))))...-.......... ( -31.40) >DroYak_CAF1 1159 113 - 1 -ACU--ACAACAGCUGCGGUAGAUAAUU-UCAAUUUCGAUUGCUGCCGGAACU-CGAUCGAAUAUUGAGAAUAUCUACCCGUACCAAUGACUCGAAAACAGCAGCAAC-CACAACAGAC -...--......(((((((((((((.((-(((((((((((((..(......).-)))))))).))))))).))))))))(((....)))...........)))))...-.......... ( -34.00) >DroAna_CAF1 1288 97 - 1 -ACC--AACAGA-ACAUGGUAGAUAAUU-UCAAUUUCGAUUGCUGCGGGAUCUUUGAUCGAAUAUUGAGAAUAUCUACCCGUAC-AAUGAAUCGA----------------CAGCAGAC -...--..((..-....((((((((.((-(((((((((((((..(......)..)))))))).))))))).)))))))).....-..))......----------------........ ( -22.72) >DroPer_CAF1 1275 103 - 1 AACUCAAGAACAGCCCUGGUAGAUAAUUUUCAAUUUCGAUUGCUGGGCGAACA-CAAUCGAUUAUUGAGAAUACCUAUC----------ACCUGACU-----AUCAACCCACAACAGAC ..........(((...((((((...(((((((((.(((((((.((......))-)))))))..)))))))))..)))))----------).)))...-----................. ( -24.30) >consensus _ACU__ACAACAGCCGUGGUAGAUAAUU_UCAAUUUCGAUUGCUGCCGGAACU_CGAUCGAAUAUUGAGAAUAUCUACCCGUAC_AAUGACUCGAAU_____AGCAAC_CACAACAGAC .................((((((((.((.(((((((((((((............)))))))).))))).))))))))))........................................ (-17.09 = -17.03 + -0.05)

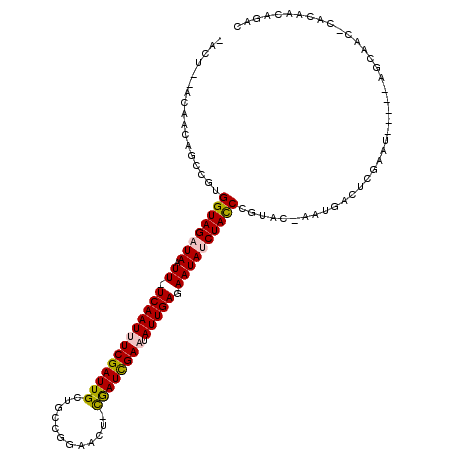

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:51 2006