
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,338,128 – 16,338,246 |
| Length | 118 |
| Max. P | 0.934509 |

| Location | 16,338,128 – 16,338,246 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -54.26 |
| Consensus MFE | -33.70 |
| Energy contribution | -33.98 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
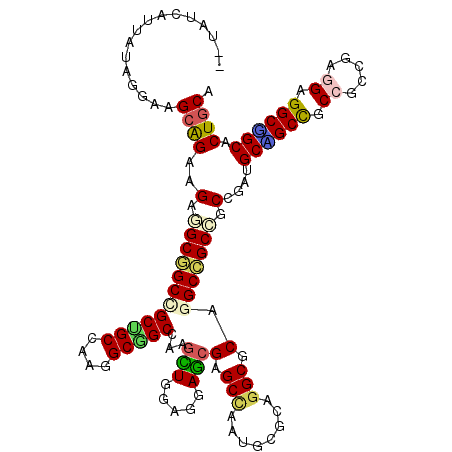
>X_DroMel_CAF1 16338128 118 + 22224390 UGCAGUGCAGCCUCUUCGGCGGCAGCCGCAUCCGCUGCUGCCUGAGCCUGCGCAUUGGCCCGCUCCUCCAGCUUGGCCGCUUUGGCGGCGGCAGCUUCUUCCGCUUCCUACAACAAUU-- .((.((((((.(((...(((((((((.......))))))))).))).))))))...((((.(((.....)))..))))))...((.(((((.((...)).))))).))..........-- ( -53.10) >DroVir_CAF1 131 118 + 1 UGCAGUGCCGCCUCUUCGGCGGCAGCGGCAUCGGCGGCGGCCUGUGCAUGCGCAUUGGCGCGCUCCUCCAGCUUAGCCGCCUUCGCAGCGGCCGCCUCCUCUGCCUCCUAUAAUGAUA-- .((..(((((((.....)))))))))((((..((.(((((((...((.((((....((((.(((..........)))))))..))))))))))))).))..)))).............-- ( -58.10) >DroGri_CAF1 131 118 + 1 UGCAGUGCAGCCUCCUCGGCGGCGGCUGCAUCAGCGGCGGCCUGCGCCUGCGCAUUAGCGCGCUCCUCCAACUUGGCUGCUUUCGCAGCGGCCGCCUCUUCUGCCUCCUACAAUGAUA-- .(((((((((((.((.....)).)))))))..((.(((((((.((((..((......))))))............(((((....)))))))))))).)).))))..............-- ( -54.60) >DroWil_CAF1 131 118 + 1 UGAAGAGCUGCCUCCUCGGCGGCAGCAGCAUCGGCAGCGGCCUGUGCCUGGGCAUUGGCUCUUUCCUCUAAUUUUGCUGCCUUGGCAGCUGCCGCUUCUUCUGCUUCCUAUAAUAAUG-- .(((((((.(((.....)))((((((.((...((((((((((.((((....)))).))))...............))))))...)).)))))))).))))).................-- ( -49.86) >DroMoj_CAF1 131 120 + 1 UGCAGGGCCGCCUCCUCGGCGGCGGCUGCAUCUGCGGCGGCUUGCGCCUGCGCAUUGGCUCGCUCCUCCAACUUAGCGGCCUUCGCAGCGGCCGCCUCUUCUGCCUCCUAUAAUGAUAUA .(((((((((......))))(((((((((....((((((.....)))).))((...(((.((((..........)))))))...)).)))))))))...)))))................ ( -53.50) >DroAna_CAF1 131 118 + 1 UGCAGGGCGGCCUCCUCGGCGGCGGCCGCUUCCGCCGCUGCCUGCGCCUGCGCAUUGGCCCGCUCCUCCAGCUUGGCCGCCUUGGCGGCAGCUGCCUCCUCCGCUUCCUACAACGAUC-- .(((((((((((.....(((((((((.......)))))))))((((....))))..)).)))).))).(((((..(((((....))))))))))........))..............-- ( -56.40) >consensus UGCAGUGCAGCCUCCUCGGCGGCAGCCGCAUCCGCGGCGGCCUGCGCCUGCGCAUUGGCCCGCUCCUCCAACUUGGCCGCCUUCGCAGCGGCCGCCUCUUCUGCCUCCUACAAUGAUA__ .........(((.....)))(((((..((....))(((((((((((....)))).....................(((((....))))))))))))....)))))............... (-33.70 = -33.98 + 0.28)


| Location | 16,338,128 – 16,338,246 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -58.22 |
| Consensus MFE | -43.67 |
| Energy contribution | -43.40 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.44 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934509 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16338128 118 - 22224390 --AAUUGUUGUAGGAAGCGGAAGAAGCUGCCGCCGCCAAAGCGGCCAAGCUGGAGGAGCGGGCCAAUGCGCAGGCUCAGGCAGCAGCGGAUGCGGCUGCCGCCGAAGAGGCUGCACUGCA --..(((((......))))).....(((((((((((....)))))...(((.....)))(((((........))))).)))))).((((.(((((((..(......).))))))))))). ( -51.50) >DroVir_CAF1 131 118 - 1 --UAUCAUUAUAGGAGGCAGAGGAGGCGGCCGCUGCGAAGGCGGCUAAGCUGGAGGAGCGCGCCAAUGCGCAUGCACAGGCCGCCGCCGAUGCCGCUGCCGCCGAAGAGGCGGCACUGCA --.............((((..((.((((((((((((....)))))...(((.....)))((((....)))).......))))))).))..))))(((((((((.....)))))))..)). ( -66.10) >DroGri_CAF1 131 118 - 1 --UAUCAUUGUAGGAGGCAGAAGAGGCGGCCGCUGCGAAAGCAGCCAAGUUGGAGGAGCGCGCUAAUGCGCAGGCGCAGGCCGCCGCUGAUGCAGCCGCCGCCGAGGAGGCUGCACUGCA --..............((((.((.((((((((((((....)))(((..(((.....)))((((....)))).))))).))))))).))..(((((((.((.....)).))))))))))). ( -62.80) >DroWil_CAF1 131 118 - 1 --CAUUAUUAUAGGAAGCAGAAGAAGCGGCAGCUGCCAAGGCAGCAAAAUUAGAGGAAAGAGCCAAUGCCCAGGCACAGGCCGCUGCCGAUGCUGCUGCCGCCGAGGAGGCAGCUCUUCA --.................(((((.((((((((.((...((((((....((....))..(.(((..(((....)))..))))))))))...)).))))))(((.....))).))))))). ( -50.10) >DroMoj_CAF1 131 120 - 1 UAUAUCAUUAUAGGAGGCAGAAGAGGCGGCCGCUGCGAAGGCCGCUAAGUUGGAGGAGCGAGCCAAUGCGCAGGCGCAAGCCGCCGCAGAUGCAGCCGCCGCCGAGGAGGCGGCCCUGCA ..........((((.(((......(((((((((((((..(((((((..........)))).)))....)))).)))...))))))((....)).)))((((((.....)))))))))).. ( -57.30) >DroAna_CAF1 131 118 - 1 --GAUCGUUGUAGGAAGCGGAGGAGGCAGCUGCCGCCAAGGCGGCCAAGCUGGAGGAGCGGGCCAAUGCGCAGGCGCAGGCAGCGGCGGAAGCGGCCGCCGCCGAGGAGGCCGCCCUGCA --......((((((..((((....(((.((((((((....))((((..(((.....))).))))..((((....))))))))))(((((......)))))))).......)))))))))) ( -61.50) >consensus __UAUCAUUAUAGGAAGCAGAAGAGGCGGCCGCUGCCAAGGCGGCCAAGCUGGAGGAGCGAGCCAAUGCGCAGGCGCAGGCCGCCGCCGAUGCAGCCGCCGCCGAGGAGGCGGCACUGCA ................((((..(.((((((((((((....)))))...(((.....)))(.(((........))).).))))))).)....((((((.((.....)).)))))).)))). (-43.67 = -43.40 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:50 2006