
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,316,877 – 16,317,037 |
| Length | 160 |
| Max. P | 0.999928 |

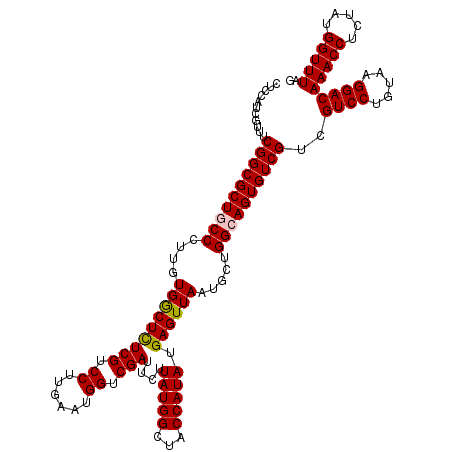
| Location | 16,316,877 – 16,316,997 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.11 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -40.18 |
| Energy contribution | -40.15 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.21 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.61 |
| SVM RNA-class probability | 0.999928 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16316877 120 + 22224390 CUCCAUCGUCUCGGCGCUGCCCUUGUGGCUCUCGUCCUUGAAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAG ...........((((((((((....(((((((((.((......)).)))....(((((...))))).)))))).....))))))))))..((((.....))))(((((.....))))).. ( -42.70) >DroVir_CAF1 23393 120 + 1 CUCCAUCGUUUCGGCGCUACCCUUGUGGCUUUCGUCCUUGAAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAG ...............(((((....)))))....(((((((...((.((((...(((.((((.(((........))).)))).)))..)))).))..)))))))(((((.....))))).. ( -34.30) >DroGri_CAF1 22509 120 + 1 UUCCAUCGUUUCGGCGCUACCCUUGUGGCUUUCGUCCUUGAAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAG ...............(((((....)))))....(((((((...((.((((...(((.((((.(((........))).)))).)))..)))).))..)))))))(((((.....))))).. ( -34.30) >DroWil_CAF1 24813 120 + 1 CUCCAUCGUUUCGGCGCUGCCCUUGUGGCUCUCGUCCUUGAAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAG ...........((((((((((....(((((((((.((......)).)))....(((((...))))).)))))).....))))))))))..((((.....))))(((((.....))))).. ( -42.70) >DroYak_CAF1 18560 120 + 1 CUCCAUCGUCUCGGCGCUGCCCUUGUGGCUCUCGUCCUUGAAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAG ...........((((((((((....(((((((((.((......)).)))....(((((...))))).)))))).....))))))))))..((((.....))))(((((.....))))).. ( -42.70) >DroMoj_CAF1 23309 120 + 1 CUCCAUCGUUUCGGCGCUGCCCUUGUGACUCUCGUCCUUGAAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAG ...........((((((((((....(((((((((.((......)).)))....(((((...))))).)))))).....))))))))))..((((.....))))(((((.....))))).. ( -43.20) >consensus CUCCAUCGUUUCGGCGCUGCCCUUGUGGCUCUCGUCCUUGAAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAG ...........((((((((((....(((((((((.((......)).)))....(((((...))))).)))))).....))))))))))..((((.....))))(((((.....))))).. (-40.18 = -40.15 + -0.03)


| Location | 16,316,877 – 16,316,997 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.11 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -30.51 |
| Energy contribution | -30.07 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16316877 120 - 22224390 CUAAACCAUAGAGGUUUGUCCUUACAGGACGACGACACUGCCAGCAUUAACUCAUAUGGUAGCCAUAAGAAUCGACCAUUCAAGGACGAGAGCCACAAGGGCAGCGCCGAGACGAUGGAG .....((((...(..((((((.....))))))((.(.(((((........(((.(((((...)))))....(((.((......)).)))))).......))))).).))...).)))).. ( -32.06) >DroVir_CAF1 23393 120 - 1 CUAAACCAUAGAGGUUUGUCCUUACAGGACGACGACACUGCCAGCAUUAACUCAUAUGGUAGCCAUAAGAAUCGACCAUUCAAGGACGAAAGCCACAAGGGUAGCGCCGAAACGAUGGAG .....((((...(..((((((.....))))))((.(.(((((.........((.(((((...))))).)).............((.(....))).....))))).).))...).)))).. ( -29.20) >DroGri_CAF1 22509 120 - 1 CUAAACCAUAGAGGUUUGUCCUUACAGGACGACGACACUGCCAGCAUUAACUCAUAUGGUAGCCAUAAGAAUCGACCAUUCAAGGACGAAAGCCACAAGGGUAGCGCCGAAACGAUGGAA .....((((...(..((((((.....))))))((.(.(((((.........((.(((((...))))).)).............((.(....))).....))))).).))...).)))).. ( -29.20) >DroWil_CAF1 24813 120 - 1 CUAAACCAUAGAGGUUUGUCCUUACAGGACGACGACACUGCCAGCAUUAACUCAUAUGGUAGCCAUAAGAAUCGACCAUUCAAGGACGAGAGCCACAAGGGCAGCGCCGAAACGAUGGAG .....((((...(..((((((.....))))))((.(.(((((........(((.(((((...)))))....(((.((......)).)))))).......))))).).))...).)))).. ( -32.06) >DroYak_CAF1 18560 120 - 1 CUAAACCAUAGAGGUUUGUCCUUACAGGACGACGACACUGCCAGCAUUAACUCAUAUGGUAGCCAUAAGAAUCGACCAUUCAAGGACGAGAGCCACAAGGGCAGCGCCGAGACGAUGGAG .....((((...(..((((((.....))))))((.(.(((((........(((.(((((...)))))....(((.((......)).)))))).......))))).).))...).)))).. ( -32.06) >DroMoj_CAF1 23309 120 - 1 CUAAACCAUAGAGGUUUGUCCUUACAGGACGACGACACUGCCAGCAUUAACUCAUAUGGUAGCCAUAAGAAUCGACCAUUCAAGGACGAGAGUCACAAGGGCAGCGCCGAAACGAUGGAG .....((((...(..((((((.....))))))((.(.(((((.......((((.(((((...)))))....(((.((......)).)))))))......))))).).))...).)))).. ( -33.72) >consensus CUAAACCAUAGAGGUUUGUCCUUACAGGACGACGACACUGCCAGCAUUAACUCAUAUGGUAGCCAUAAGAAUCGACCAUUCAAGGACGAGAGCCACAAGGGCAGCGCCGAAACGAUGGAG .....((((....((((((((.....))))..((.(.(((((.........((.(((((...))))).)).(((.((......)).)))..........))))).).)))))).)))).. (-30.51 = -30.07 + -0.44)
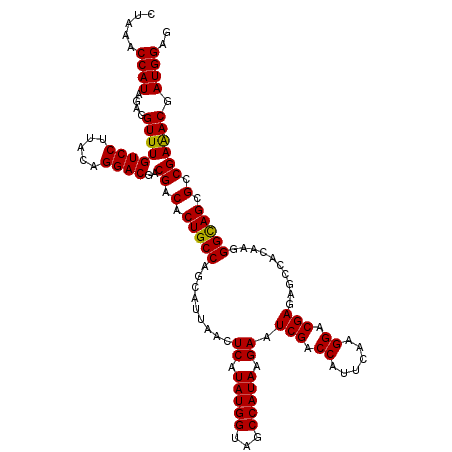

| Location | 16,316,917 – 16,317,037 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -34.10 |
| Energy contribution | -34.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16316917 120 + 22224390 AAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAGCUCGUGUUCCAGUCUAUUGUCUUGGUGGUUAAUUGAGUUG ......((((((...(((((((((((((((((......(((......)))((((.....))))(((((.....))))))))))))))...((.(....).)).))))))))...)))))) ( -34.10) >DroEre_CAF1 19023 120 + 1 AAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAGCUCGUGUUCCAGUCUAUUGUCUUGGUGGUUAAUUGAGUUG ......((((((...(((((((((((((((((......(((......)))((((.....))))(((((.....))))))))))))))...((.(....).)).))))))))...)))))) ( -34.10) >DroWil_CAF1 24853 120 + 1 AAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAGCUCGUGUUCCAGUCUAUUGUCUUGGUGGUUAAUUGAGUUG ......((((((...(((((((((((((((((......(((......)))((((.....))))(((((.....))))))))))))))...((.(....).)).))))))))...)))))) ( -34.10) >DroYak_CAF1 18600 120 + 1 AAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAGCUCGUGUUCCAGUCUAUUGUCUUGGUGGUUAAUUGAGUUG ......((((((...(((((((((((((((((......(((......)))((((.....))))(((((.....))))))))))))))...((.(....).)).))))))))...)))))) ( -34.10) >DroMoj_CAF1 23349 120 + 1 AAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAGCUCGUGUUCCAGUCUAUUGUCUUGGUGGUUAAUUGAGUUG ......((((((...(((((((((((((((((......(((......)))((((.....))))(((((.....))))))))))))))...((.(....).)).))))))))...)))))) ( -34.10) >DroAna_CAF1 18630 120 + 1 AAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAGCUCGUGUUCCAGUCUAUUGUCUUGGUGGUUAAUUGAGUUG ......((((((...(((((((((((((((((......(((......)))((((.....))))(((((.....))))))))))))))...((.(....).)).))))))))...)))))) ( -34.10) >consensus AAUGGUCGAUUCUUAUGGCUACCAUAUGAGUUAAUGCUGGCAGUGUCGUCGUCCUGUAAGGACAAACCUCUAUGGUUUAGCUCGUGUUCCAGUCUAUUGUCUUGGUGGUUAAUUGAGUUG ......((((((...(((((((((((((((((......(((......)))((((.....))))(((((.....))))))))))))))...((.(....).)).))))))))...)))))) (-34.10 = -34.10 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:41 2006