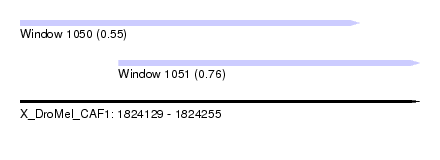
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,824,129 – 1,824,255 |
| Length | 126 |
| Max. P | 0.762441 |

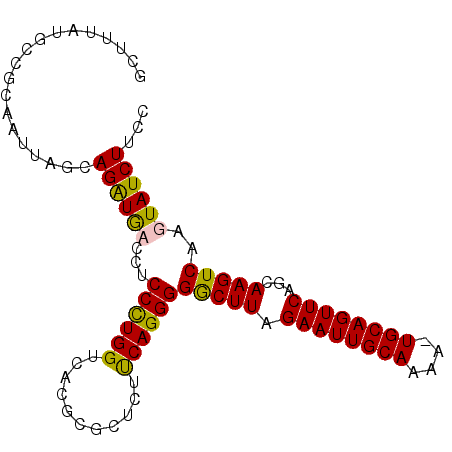
| Location | 1,824,129 – 1,824,236 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.08 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -28.27 |
| Energy contribution | -28.02 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545985 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1824129 107 + 22224390 CGCCUUGGUGAAUGUGGUAAGUGUCGGGAUUACUUUGUGCCACAAUUAGCAGAUGCCCUCCUUGGUCACGCGCUCACCAGGGGGCUUAGAAUUGCAAAA-UGCAGUUC (.(((((((((.(((((((((((.......))))...)))))))....((.(.((.((.....)).)))..))))))))))).)....(((((((....-.))))))) ( -37.30) >DroSec_CAF1 24741 107 + 1 CGCCUUGGUGAAUGUGGUAAGUGUCGGAAUUGCUUUAUGCCGCAAUUAGCAGAUGAUCUCCCUGGUCACGCGCUCUUCAGGGGGCUUAGAAUUGCAAAA-UGCAGUUC .((((((.(((.(((((((((((.......))))...))))))).))).))).......((((((...........)))))))))...(((((((....-.))))))) ( -33.00) >DroSim_CAF1 26653 107 + 1 CGCCUUGGUGAAUGUGGUAAGUGUCGGAAUUGCUUUAUGUCGCAAUUACCAGAUGAUCUCCCUGGUCACUCGCUCUUCAGGGGGCUUAGAAUUGCAAAA-UGCAGUUC .((((((((((.(((((.(((((.......)))))....))))).))))))).......((((((...........)))))))))...(((((((....-.))))))) ( -33.30) >DroEre_CAF1 24357 108 + 1 CGCCUUGGUGAAUGUGGUAAGUGUCGGGACGGCUUUAUGCCGCAAUUAGGAGGUACCCUCCUUGGUCUCGCGCUCUUCAGGGGACUUAGAAUUGCAAAAUUGCAGUUC (((....)))....(((..((((.(((((((((.....)))......((((((...))))))..))))))))))..)))(....)...((((((((....)))))))) ( -39.90) >consensus CGCCUUGGUGAAUGUGGUAAGUGUCGGAAUUGCUUUAUGCCGCAAUUAGCAGAUGACCUCCCUGGUCACGCGCUCUUCAGGGGGCUUAGAAUUGCAAAA_UGCAGUUC .((((((.(((.(((((((((((.......))))...))))))).))).))).......((((((...........)))))))))...((((((((....)))))))) (-28.27 = -28.02 + -0.25)

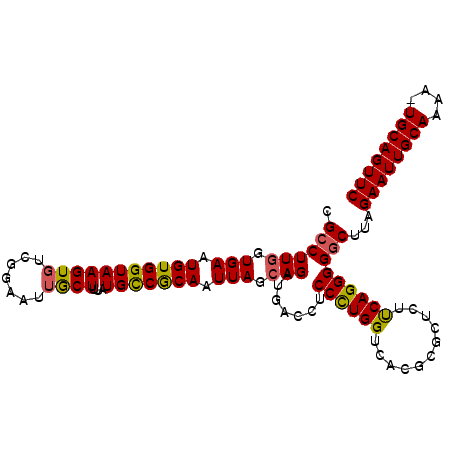
| Location | 1,824,160 – 1,824,255 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 89.53 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -24.38 |
| Energy contribution | -23.88 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1824160 95 + 22224390 ACUUUGUGCCACAAUUAGCAGAUGCCCUCCUUGGUCACGCGCUCACCAGGGGGCUUAGAAUUGCAAAA-UGCAGUUCAGCAAGUCAAGUAUCUUCC ......(((........)))(((((...(((((((.........)))))))(((((.(((((((....-.)))))))...)))))..))))).... ( -27.20) >DroSec_CAF1 24772 95 + 1 GCUUUAUGCCGCAAUUAGCAGAUGAUCUCCCUGGUCACGCGCUCUUCAGGGGGCUUAGAAUUGCAAAA-UGCAGUUCAGCAAGUCAAGUAUCUUCC (((....(((((.....)).........((((((...........)))))))))...(((((((....-.))))))))))................ ( -25.90) >DroSim_CAF1 26684 95 + 1 GCUUUAUGUCGCAAUUACCAGAUGAUCUCCCUGGUCACUCGCUCUUCAGGGGGCUUAGAAUUGCAAAA-UGCAGUUCAGCAAGUCAAGUAUCUUCC ((((.((...((..........(((.((((((((...........)))))))).)))(((((((....-.))))))).))..)).))))....... ( -25.30) >DroEre_CAF1 24388 96 + 1 GCUUUAUGCCGCAAUUAGGAGGUACCCUCCUUGGUCUCGCGCUCUUCAGGGGACUUAGAAUUGCAAAAUUGCAGUUCAUGAAGUCAAGUAUCUUCC ((........)).....((((((((...((((((...........))))))(((((.((((((((....))))))))...)))))..)))))))). ( -29.70) >consensus GCUUUAUGCCGCAAUUAGCAGAUGACCUCCCUGGUCACGCGCUCUUCAGGGGGCUUAGAAUUGCAAAA_UGCAGUUCAGCAAGUCAAGUAUCUUCC ...................((((((...((((((...........))))))(((((.((((((((....))))))))...)))))..))))))... (-24.38 = -23.88 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:19 2006