
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,315,735 – 16,315,842 |
| Length | 107 |
| Max. P | 0.940497 |

| Location | 16,315,735 – 16,315,842 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.93 |
| Mean single sequence MFE | -22.43 |
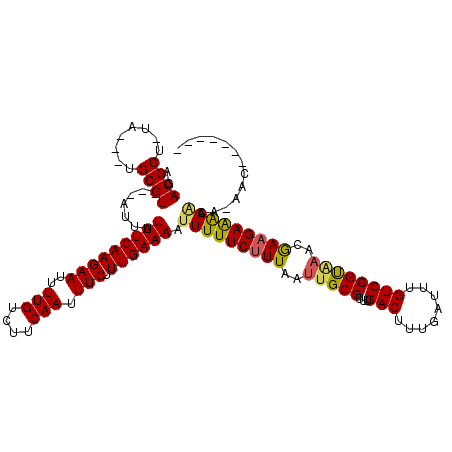
| Consensus MFE | -21.25 |
| Energy contribution | -20.92 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
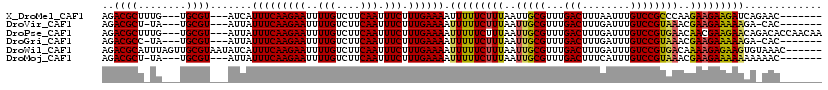
>X_DroMel_CAF1 16315735 107 - 22224390 AGACGCUUUG---UGCGU---AUCAUUUCAAGAAUUUUGUCUUCAAUUUCUUUGAAAAUUUUUCUUUAAUUGCGUUUGACUUUAAUUUGUCCGCCCAAGAAGAAGAUCAGAAC------- ..((((....---.))))---.((.(((((((((..(((....))).))).))))))..((((((((....(((...(((........))))))....))))))))...))..------- ( -21.30) >DroVir_CAF1 22210 105 - 1 AGACGCU-UA---UGCGU---AUUAUUUCAAGAAUUUUGUCUUCAAUUUCUUUGAAAAUUUUUCUUUAAUUGCGUUUGACUUUGAUUUGUCCGUAAACGAAGAAAAAGA-CAC------- ..((((.-..---.))))---................(((((((((.....)))))..(((((((((..(((((...(((........))))))))..)))))))))))-)).------- ( -23.60) >DroPse_CAF1 20781 114 - 1 AGACGCUUUG---UGCGU---AUUAUUUCAAGAAUUUUGUCUUCAAUUUCUUUGAAAAUUUUUCUUUAAUUGCGUUUGACUUUGAUUUGUCCGUGAACAACGAAGAACAGACACCAACAA ..((((....---.))))---....(((((((((..(((....))).))).))))))................(((((.(((((..((((......)))))))))..)))))........ ( -18.80) >DroGri_CAF1 21317 105 - 1 AGACGCC-UA---UGCGU---AUUAUUUCAAGAAUUUUGUCUUCAAUUUCUUUGAAAAUUUUUCUUUAAUUGCGUUUGACUUUGAUUUGUCCGUAAACGAAGAAAAAGA-CAC------- ..((((.-..---.))))---................(((((((((.....)))))..(((((((((..(((((...(((........))))))))..)))))))))))-)).------- ( -22.40) >DroWil_CAF1 23616 114 - 1 AGACGCAUUUAGUUGCGUAAUAUCAUUUCAAGAAUUUUGUCUUCAAUUUCUUUGAAAAUUUUUCUUUAAUUGCGUUUGACUUUGAUUUGUCCGUGACAAAAGAGAAGUGUAAAC------ ..(((((......))))).......(((((((((..(((....))).))).))))))((((((((((..(..((...(((........)))))..)..))))))))))......------ ( -25.20) >DroMoj_CAF1 22151 106 - 1 AGACGCU-UA---UGCGU---AUUAUUUCAAGAAUUUUGUCUUCAAUUUCUUUGAAAAUUUUUCUUUAAUUGCGUUUGACUUUCAUUUGUCCGUAAACGAAGAAAAAAAAAAC------- ..((((.-..---.))))---....(((((((((..(((....))).))).)))))).(((((((((..(((((...(((........))))))))..)))))))))......------- ( -23.30) >consensus AGACGCU_UA___UGCGU___AUUAUUUCAAGAAUUUUGUCUUCAAUUUCUUUGAAAAUUUUUCUUUAAUUGCGUUUGACUUUGAUUUGUCCGUAAACGAAGAAAAACA_AAC_______ ..((((........)))).......(((((((((..(((....))).))).)))))).(((((((((..(((((...(((........))))))))..)))))))))............. (-21.25 = -20.92 + -0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:37 2006