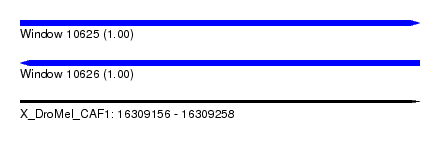
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,309,156 – 16,309,258 |
| Length | 102 |
| Max. P | 0.999135 |

| Location | 16,309,156 – 16,309,258 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 70.55 |
| Mean single sequence MFE | -24.61 |
| Consensus MFE | -14.63 |
| Energy contribution | -14.99 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
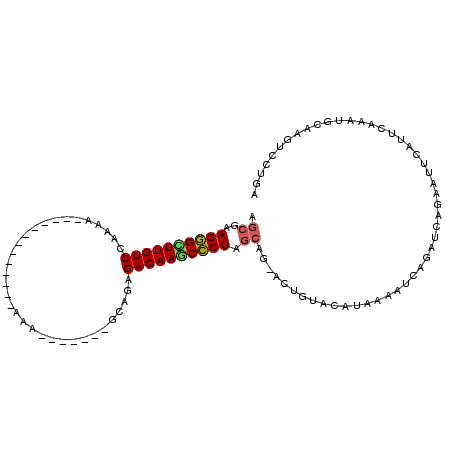
>X_DroMel_CAF1 16309156 102 + 22224390 UCAGGACUUGCACUUGAAUGAAUUCUGAUUUGAUUUUAUGUACAGUACAGCUAGGGCUUGUCUCUGA-------UUUUUUUUUAU-GUUUUUUUGGACAAGCCCUUUGCU (((((.......)))))(((((.((......)).))))).........(((.(((((((((((..((-------...........-....))..)))))))))))..))) ( -26.36) >DroPse_CAF1 13418 94 + 1 --------------UGAAUGAAUUC-GAUUCGAUAUUAUAUACAAU-CUGCAAGAGUUUGUCUUCGGAGGUCGGUCUCCCUUGGUAGAUCGGUUCGACAAACUCUUGUUU --------------.((((......-.))))...............-..(((((((((((((...(((..(((((((((...)).))))))))))))))))))))))).. ( -26.80) >DroSec_CAF1 10861 90 + 1 UCAGGACUUGCACUUGAAUGAAUUCUGAUCUGAUUUUAUGUACAGU-CAGCUAGGGGUUGUCUUUGA-------UUUU------------UUUUGGACAAGCCCUUCGCU ....((((((((......((((.((......)).)))))))).)))-)(((.((((.((((((..((-------....------------))..)))))).))))..))) ( -24.60) >DroEre_CAF1 11343 89 + 1 UCAGGACUUGCAUUUGAAUGAAUUCUGAUCUGAUUUUAUGUACAUU-CUGCUAGGGCUUGUCUCUGC-------UUU-------------CUUUGGACAAACCCUUCGCU ..((((..(((((..((((.(.........).)))).)))))..))-))((.((((.((((((....-------...-------------....)))))).))))..)). ( -17.80) >DroYak_CAF1 10686 89 + 1 UCAGGACUUGCAUUGGAAUGAAUUCUGAUCUGAUAUUAUGUACAGU-CUGCUAGGGCUUGUCUCUGC-------UUU-------------UUAUGGACAAGCCCUUCGCU ...((((((((((..(((....)))..)..........)))).)))-))((.(((((((((((.((.-------...-------------.)).)))))))))))..)). ( -27.50) >consensus UCAGGACUUGCACUUGAAUGAAUUCUGAUCUGAUUUUAUGUACAGU_CUGCUAGGGCUUGUCUCUGA_______UUU_____________UUUUGGACAAGCCCUUCGCU .................(((((.((......)).)))))..........((.((((((((((.................................))))))))))..)). (-14.63 = -14.99 + 0.36)



| Location | 16,309,156 – 16,309,258 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 70.55 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -13.89 |
| Energy contribution | -13.89 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.999135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16309156 102 - 22224390 AGCAAAGGGCUUGUCCAAAAAAAC-AUAAAAAAAAA-------UCAGAGACAAGCCCUAGCUGUACUGUACAUAAAAUCAAAUCAGAAUUCAUUCAAGUGCAAGUCCUGA (((..(((((((((((........-...........-------...).)))))))))).)))(((((..........((......)).........)))))......... ( -21.96) >DroPse_CAF1 13418 94 - 1 AAACAAGAGUUUGUCGAACCGAUCUACCAAGGGAGACCGACCUCCGAAGACAAACUCUUGCAG-AUUGUAUAUAAUAUCGAAUC-GAAUUCAUUCA-------------- ...((((((((((((((.....)).......((((......))))...))))))))))))...-...............((((.-(....))))).-------------- ( -23.90) >DroSec_CAF1 10861 90 - 1 AGCGAAGGGCUUGUCCAAAA------------AAAA-------UCAAAGACAACCCCUAGCUG-ACUGUACAUAAAAUCAGAUCAGAAUUCAUUCAAGUGCAAGUCCUGA (((..((((.(((((.....------------....-------.....))))).)))).)))(-(((((((..........................)))).)))).... ( -18.71) >DroEre_CAF1 11343 89 - 1 AGCGAAGGGUUUGUCCAAAG-------------AAA-------GCAGAGACAAGCCCUAGCAG-AAUGUACAUAAAAUCAGAUCAGAAUUCAUUCAAAUGCAAGUCCUGA .((..(((((((((((....-------------...-------...).)))))))))).)).(-((((......................)))))............... ( -19.95) >DroYak_CAF1 10686 89 - 1 AGCGAAGGGCUUGUCCAUAA-------------AAA-------GCAGAGACAAGCCCUAGCAG-ACUGUACAUAAUAUCAGAUCAGAAUUCAUUCCAAUGCAAGUCCUGA .((..(((((((((((....-------------...-------...).)))))))))).))..-.(((..........))).((((.((((((....)))..))).)))) ( -22.50) >consensus AGCGAAGGGCUUGUCCAAAA_____________AAA_______GCAGAGACAAGCCCUAGCAG_ACUGUACAUAAAAUCAGAUCAGAAUUCAUUCAAAUGCAAGUCCUGA .((..((((((((((.................................)))))))))).))................................................. (-13.89 = -13.89 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:32 2006