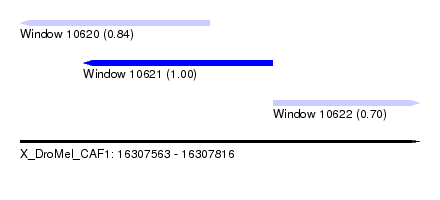
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,307,563 – 16,307,816 |
| Length | 253 |
| Max. P | 0.997059 |

| Location | 16,307,563 – 16,307,683 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -20.40 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.86 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16307563 120 - 22224390 ACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUUCAUGACAGAAGAUCAGAAAAAGUACUAUAAUGCUAUGAAA ....(((...((((((((((.((((((...)))))).)))))))))).........((((.((((.((....))....(((.((((....).))))))......))))..)))).))).. ( -20.40) >DroEre_CAF1 9775 120 - 1 ACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUUCAUGACAGAAGAUCAGAAAAAGUACUAUAAUGCUAUGAAA ....(((...((((((((((.((((((...)))))).)))))))))).........((((.((((.((....))....(((.((((....).))))))......))))..)))).))).. ( -20.40) >DroWil_CAF1 12738 120 - 1 ACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUUCAUGACAGAAGAUCAGAAAAAGUACUAUAAUGCUAUGAAA ....(((...((((((((((.((((((...)))))).)))))))))).........((((.((((.((....))....(((.((((....).))))))......))))..)))).))).. ( -20.40) >DroYak_CAF1 9085 120 - 1 ACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUUCAUGACAGAAGAUCAGAAAAAGUACUAUAAUGCUAUGAAA ....(((...((((((((((.((((((...)))))).)))))))))).........((((.((((.((....))....(((.((((....).))))))......))))..)))).))).. ( -20.40) >DroMoj_CAF1 11504 120 - 1 ACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUUCAUGACAGAAGAUCAGAAAAAGUACUAUAAUGCUAUGAAA ....(((...((((((((((.((((((...)))))).)))))))))).........((((.((((.((....))....(((.((((....).))))))......))))..)))).))).. ( -20.40) >DroAna_CAF1 9579 120 - 1 ACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUUCAUGACAGAAGAUCAGAAAAAGUACUAUAAUGCUAUGAAA ....(((...((((((((((.((((((...)))))).)))))))))).........((((.((((.((....))....(((.((((....).))))))......))))..)))).))).. ( -20.40) >consensus ACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUUCAUGACAGAAGAUCAGAAAAAGUACUAUAAUGCUAUGAAA ....(((...((((((((((.((((((...)))))).)))))))))).........((((.((((.((....))....(((.((((....).))))))......))))..)))).))).. (-20.40 = -20.40 + 0.00)



| Location | 16,307,603 – 16,307,723 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.50 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -24.40 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16307603 120 - 22224390 UUUAUAUUUCGUAUUCUUCAUCAUAUUUGGAUCAUUUUUCACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUU ...(((((((......((((((.....((((......)))).........((((((((((.((((((...)))))).)))))))))).............))))))......))))))). ( -24.40) >DroVir_CAF1 10968 120 - 1 UUUAUAUUUCGUGUUCUUCAUCAUAUUUGGAUCAUUUUUCACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUU ...(((((((......((((((.....((((......)))).........((((((((((.((((((...)))))).)))))))))).............))))))......))))))). ( -24.40) >DroGri_CAF1 11508 120 - 1 UUUAUAUUUCGUGUUCUUCAUCAUAUUUGGAUCAUUUUUCACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUU ...(((((((......((((((.....((((......)))).........((((((((((.((((((...)))))).)))))))))).............))))))......))))))). ( -24.40) >DroYak_CAF1 9125 120 - 1 UUUAUAUUUCGUAUUCUUCAUCAUAUUUGGAUCAUUUUUCACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUU ...(((((((......((((((.....((((......)))).........((((((((((.((((((...)))))).)))))))))).............))))))......))))))). ( -24.40) >DroMoj_CAF1 11544 120 - 1 UUUAUAUUUCGUGUUCUUCAUCAUAUUUGGAUCAUUUUUCACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUU ...(((((((......((((((.....((((......)))).........((((((((((.((((((...)))))).)))))))))).............))))))......))))))). ( -24.40) >DroAna_CAF1 9619 120 - 1 UUUAUAUUUCGUAUUCUUCAUCAUAUUUGGAUCAUUUUUCACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUU ...(((((((......((((((.....((((......)))).........((((((((((.((((((...)))))).)))))))))).............))))))......))))))). ( -24.40) >consensus UUUAUAUUUCGUAUUCUUCAUCAUAUUUGGAUCAUUUUUCACACUCAAUCUGUUCAUUGGUGUUAUCAUUGAUAAUUUUAAUGAGCAAAAGAAAAAAGCAGGUGGAUCAUUAGAAAUGUU ...(((((((......((((((.....((((......)))).........((((((((((.((((((...)))))).)))))))))).............))))))......))))))). (-24.40 = -24.40 + 0.00)

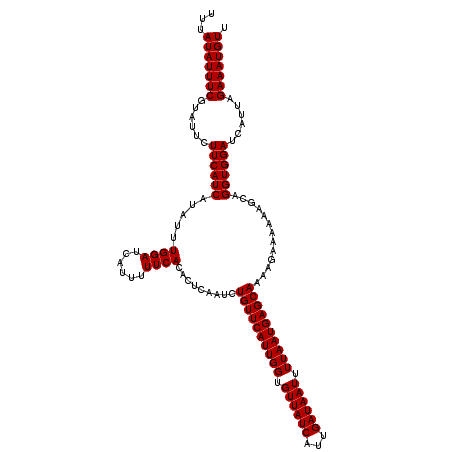
| Location | 16,307,723 – 16,307,816 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.93 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16307723 93 + 22224390 UACAUGUAGAUGUUCGUUUCACGAAUUGGUUGCUUGUCCACCUGUUAUCGAUCAUU---------UGGCCAG-----UUCGAAGUCAAUCGAUAGCAGCAGGAAUUG ..((.((((..(((((.....)))))...)))).))(((..(((((((((((..((---------(((....-----.)))))....)))))))))))..))).... ( -26.60) >DroVir_CAF1 11088 79 + 1 UACAUGUAGAUGUUCGUUUCACGAAUCGGCUGCUUGUCCACCUG-UAUCGAU-AA------CGAUA-GCGAU-----AACGA----UAUCGAUAUCA---------- .(((.((((..(((((.....)))))...)))).)))......(-(((((((-(.------((.((-....)-----).)).----)))))))))..---------- ( -20.30) >DroSec_CAF1 9448 93 + 1 UACAUGUAGAUGUUCGUUUCACGAAUUGGUUGCUUGUCCACCUGUUAUCGAUCAUU---------UGGCCAG-----UUCGAAGUCAAUCGAUAGCAGCAGGAAUCG ..((.((((..(((((.....)))))...)))).))(((..(((((((((((..((---------(((....-----.)))))....)))))))))))..))).... ( -26.60) >DroEre_CAF1 9935 93 + 1 UACAUGUAGAUGUUCGUUUCACGAAUUGGUUGCUUGUCCACCUGUUAUCGAUCAUU---------UGGCCAG-----UUCGAAGUCAAUCGAUAGCAGCAGGAAUCG ..((.((((..(((((.....)))))...)))).))(((..(((((((((((..((---------(((....-----.)))))....)))))))))))..))).... ( -26.60) >DroYak_CAF1 9245 93 + 1 UACAUGUAGAUGUUCGUUUCACGAAUUGGUUGCUUGUCCACCUGUUAUCGAUCAUU---------UGGCCAG-----UUCGAAGUCAAUCGAUAGCAGCAGGAAUCG ..((.((((..(((((.....)))))...)))).))(((..(((((((((((..((---------(((....-----.)))))....)))))))))))..))).... ( -26.60) >DroAna_CAF1 9739 103 + 1 UACAUGUAGAUGUUCGUUUCACGAAUUGGCUGCUUGUCCACCUGUUAUCGAUAAUUUAUUGCGAUUGGCCAGAUGAUUUUGA----GAUCGAUAGCAGCAGGAAUCG ..((.((((..(((((.....)))))...)))).))(((..(((((((((((........((.....))((((....)))).----.)))))))))))..))).... ( -29.50) >consensus UACAUGUAGAUGUUCGUUUCACGAAUUGGUUGCUUGUCCACCUGUUAUCGAUCAUU_________UGGCCAG_____UUCGAAGUCAAUCGAUAGCAGCAGGAAUCG ..((.((((..(((((.....)))))...)))).))(((..(((((((((((...................................)))))))))))..))).... (-20.32 = -20.93 + 0.61)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:28 2006