
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,302,660 – 16,302,759 |
| Length | 99 |
| Max. P | 0.905324 |

| Location | 16,302,660 – 16,302,759 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.14 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -20.51 |
| Energy contribution | -20.37 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
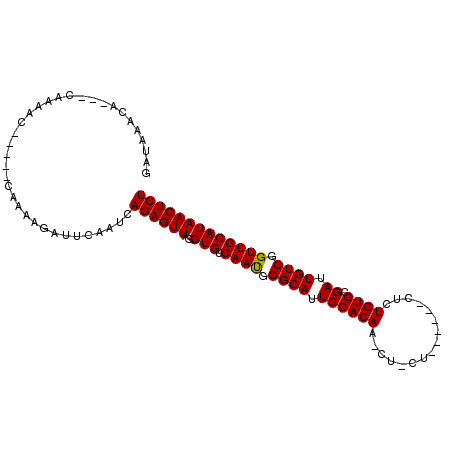
>X_DroMel_CAF1 16302660 99 - 22224390 GAUAAACU---CAAUCCAAUCACACAAGAUUCAUUCAGAGUUAGGUGUCAAUGGGCAUUCCACAA-GCAUU---CCACUCUGUGCGAUUGUCAGUUGCAUAACUCU ........---((((.(((((.(((..((.....))(((((..(((((...(((.....)))...-)))))---..)))))))).)))))...))))......... ( -20.70) >DroVir_CAF1 5786 94 - 1 GAUAAAAAGCACAACAC-----GAAAAGAUUCAAUCAGAGUUAGGUGUCAAUGGGCAUUCCACAAACU-CU------AUAUGUGCGAUUGUCGGUUGCAUAACUCU .................-----..............((((((..(((.((((.((((.((((((....-..------...)))).)).)))).))))))))))))) ( -21.30) >DroPse_CAF1 5987 92 - 1 GAAGAGGA---GAAA-AAUAUACAAAAGAUUCAAUCAGAGUUAGGUGUCAAUGGGCAUUCCACAA-A---------ACUCUGUGCGAUUGUCGGUUGCAUAACUCU ..((((..---....-...........(((((.....)))))..(((.((((.((((.((((((.-.---------....)))).)).)))).)))))))..)))) ( -23.10) >DroGri_CAF1 6246 90 - 1 GAUAAACA---CAACAC-----GAUAAGAUUCAAUCAGAGUUAGGUGUCAAUGGGCAUUCCACAA-CU-CU------CAGUGUGCGAUUGUCGGUUGCAUAACUCU ........---......-----(((........)))((((((..(((.((((.((((.((((((.-..-..------...)))).)).)))).))))))))))))) ( -22.50) >DroMoj_CAF1 5998 94 - 1 GAUAAAGAC-----GAC-----GAUAAGAUUCAAUCAGAGUUAGGUGUCAACAGGCAUUCCACAA-CU-CUCUACCGAUCUGUGCGAUUGUCGGUUGCAUAACUCU .........-----...-----(((........)))((((((..(((.((((.((((.((((((.-.(-(......))..)))).)).)))).))))))))))))) ( -23.30) >DroAna_CAF1 4828 95 - 1 GAAGAUUU---CAAAUCAAUCACACAAGAUUCAAUCAGAGUUAGGUGUCAAUGGGCAUUCCACAA-GA-------CUCUCUGUGCGAUUGUCGGUUGCAUAACUCU ...((((.---..((((..........)))).))))((((((..(((.((((.((((.((((((.-..-------.....)))).)).)))).))))))))))))) ( -23.90) >consensus GAUAAACA___CAAAAC_____CAAAAGAUUCAAUCAGAGUUAGGUGUCAAUGGGCAUUCCACAA_CU_CU______CUCUGUGCGAUUGUCGGUUGCAUAACUCU ....................................((((((..(((.((((.((((.((((((................)))).)).)))).))))))))))))) (-20.51 = -20.37 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:24 2006