| Sequence ID | X_DroMel_CAF1 |
|---|---|
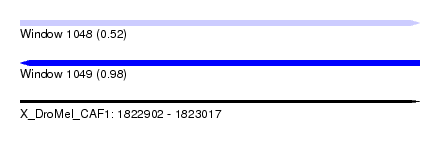
| Location | 1,822,902 – 1,823,017 |
| Length | 115 |
| Max. P | 0.982010 |

| Location | 1,822,902 – 1,823,017 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -23.57 |
| Energy contribution | -23.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
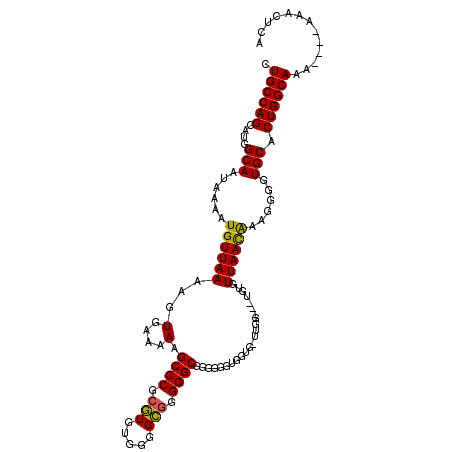
>X_DroMel_CAF1 1822902 115 + 22224390 CUGCCAGCAUGGCAAUAAAAUGUUAAAAGUGAAAAAGCCCGCGUGUGGGGCGGGGGCGGGGUGUGGUGGUUGCUGUUGUGUUAACAAAGGGGUGCACUGGCAAAAAAAAAACUCA .((((.....))))......................((((.(((.....))).)))).((((....(..(((((..((..((........))..))..)))))..)....)))). ( -31.80) >DroSec_CAF1 23659 108 + 1 CUGCCAGCAUGGCAAUAAAAUGUUAAAAGUGGAAAAGCCCGCGUGUGGGGCGGGGGCGGGGGGUGGGGCUUGG---UUUGUUAACAAAGGGGUGCACUGGCAAA----AAACUCA .((((((....(((......((((((..........((((.(((.....))).))))...(((.....)))..---....))))))......))).))))))..----....... ( -30.00) >DroEre_CAF1 23087 105 + 1 AUGCCAGCAUAGCAAUAAAAUGUUAAAAGUGAAAAAGCCCGCUUGAGGAGGAAGGGCGGGGGGAGAU---GGG---GGUGUUAAUGAAGGGGUGCACUGGCAGA----AAAUGCA .((((((..(((((......)))))............(((((((.........))))))).......---...---.(..((........))..).))))))..----....... ( -23.10) >consensus CUGCCAGCAUGGCAAUAAAAUGUUAAAAGUGAAAAAGCCCGCGUGUGGGGCGGGGGCGGGGGGUGGUG_UUGG___UGUGUUAACAAAGGGGUGCACUGGCAAA____AAACUCA .((((((....(((......((((((..........((((.(((.....))).))))..............((....)).))))))......))).))))))............. (-23.57 = -23.13 + -0.44)

| Location | 1,822,902 – 1,823,017 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -22.31 |
| Consensus MFE | -17.65 |
| Energy contribution | -17.65 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
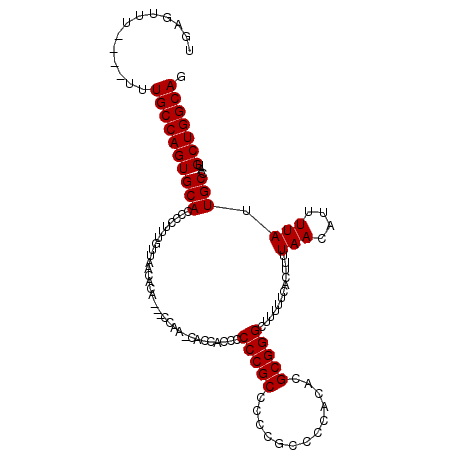
>X_DroMel_CAF1 1822902 115 - 22224390 UGAGUUUUUUUUUUGCCAGUGCACCCCUUUGUUAACACAACAGCAACCACCACACCCCGCCCCCGCCCCACACGCGGGCUUUUUCACUUUUAACAUUUUAUUGCCAUGCUGGCAG .............((((((((((......((((((.......((..............)).(((((.......)))))...........))))))......)))...))))))). ( -24.54) >DroSec_CAF1 23659 108 - 1 UGAGUUU----UUUGCCAGUGCACCCCUUUGUUAACAAA---CCAAGCCCCACCCCCCGCCCCCGCCCCACACGCGGGCUUUUCCACUUUUAACAUUUUAUUGCCAUGCUGGCAG .......----..((((((((((......((((((....---..(((((((.......((....)).......).))))))........))))))......)))...))))))). ( -24.88) >DroEre_CAF1 23087 105 - 1 UGCAUUU----UCUGCCAGUGCACCCCUUCAUUAACACC---CCC---AUCUCCCCCCGCCCUUCCUCCUCAAGCGGGCUUUUUCACUUUUAACAUUUUAUUGCUAUGCUGGCAU .......----..((((((((((................---...---.......(((((.............)))))............(((....))).)))...))))))). ( -17.52) >consensus UGAGUUU____UUUGCCAGUGCACCCCUUUGUUAACACA___CCAA_CACCACCCCCCGCCCCCGCCCCACACGCGGGCUUUUUCACUUUUAACAUUUUAUUGCCAUGCUGGCAG .............((((((((((................................(((((.............)))))............(((....))).)))...))))))). (-17.65 = -17.65 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:18 2006