
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 301,168 – 301,259 |
| Length | 91 |
| Max. P | 0.722690 |

| Location | 301,168 – 301,259 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.62 |
| Mean single sequence MFE | -17.22 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.87 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.722690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
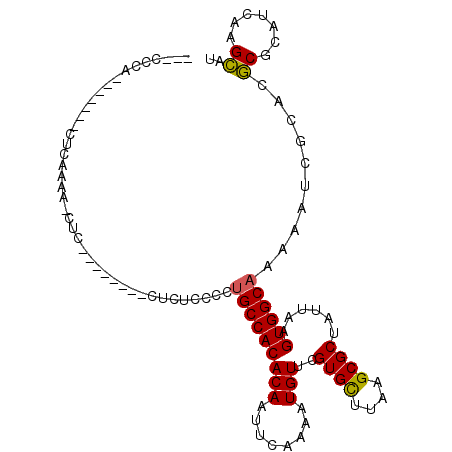
>X_DroMel_CAF1 301168 91 - 22224390 ---CCCA-------CUUAAAA-CUU--------UUCUCCCCUGCCACACAAUUCAAAAUGUUCGUGCUUAAGCGCUAUUAAGUGGCAAAAAUCGCACGCGCAUCAAGCAU ---....-------.......-...--------........(((((((((........)))..((((....))))......))))))..........((.......)).. ( -16.30) >DroSec_CAF1 44228 91 - 1 ---CCCA-------CUCAAAA-CUU--------CUCUCCCCUGCCACACAAUUCAAAAUGUUCGUGCUUAAGCGCUAUUAAGUGGCAAAAAUCGCACGCGCAUCAAGCAU ---....-------.......-...--------........(((((((((........)))..((((....))))......))))))..........((.......)).. ( -16.30) >DroSim_CAF1 64855 91 - 1 ---CCCA-------CUCAAAA-CUU--------CUCUCCCCUGCCACACAAUUCAAAAUGUUCGUGCUUAAGCGCUAUUAAGUGGCAAAAAUCGCACGCGCAUCAAGCAU ---....-------.......-...--------........(((((((((........)))..((((....))))......))))))..........((.......)).. ( -16.30) >DroEre_CAF1 43238 94 - 1 AUCCCCA-------CUCAAAA-UUC--------CUCUCCCCUGCCACACAAUUCAAAAUGUUCGUGCUUAAGCGCUAUUAAGUGGCAAAAAUCGCACGCGCAUCAAGCAU .......-------.......-...--------........(((((((((........)))..((((....))))......))))))..........((.......)).. ( -16.30) >DroYak_CAF1 65111 102 - 1 AUCCCCA-------CUCAAAA-CUCCCUGCCGCCUCUCCCCUGCCACACAAUUCAAAAUGUUCGUGCUUAAGCGCUAUUAAGUGGCAAAAAUCGCACGCGCAUCAAGCGU .......-------.......-.........((........(((((((((........)))..((((....))))......))))))......))((((.......)))) ( -19.14) >DroAna_CAF1 57821 99 - 1 ---CCCAAGGAGCCCCCAAGAUCCC--------CUCGUCCUGGCCACACAAUUCAAAAUGUUCGUGUUUAAGCGCUAUUAAGUGGCAAAAAUCGCGCUCACAUCAAGACU ---......((((...((.(((...--------...))).))((((((((........)))..((((....))))......))))).........))))........... ( -19.00) >consensus ___CCCA_______CUCAAAA_CUC________CUCUCCCCUGCCACACAAUUCAAAAUGUUCGUGCUUAAGCGCUAUUAAGUGGCAAAAAUCGCACGCGCAUCAAGCAU .........................................(((((((((........)))..((((....))))......))))))..........((.......)).. (-15.11 = -14.87 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:47 2006