| Sequence ID | X_DroMel_CAF1 |
|---|---|
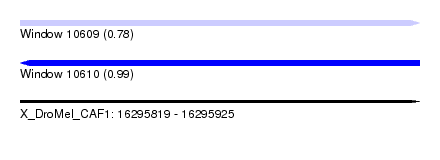
| Location | 16,295,819 – 16,295,925 |
| Length | 106 |
| Max. P | 0.987616 |

| Location | 16,295,819 – 16,295,925 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | -5.90 |
| Energy contribution | -6.26 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16295819 106 + 22224390 UGCGAA-AAUAGAGCACUUUGAAAACAAAAACUUGUUCAAAU-U-----GUUCACUCGUUUUCGUAUCUCUAUUUGC----UGUUUUUUUUUCUUU---UAUUUUGUGCUAGGCACAAAC ((((((-(((.(((((.(((((..((((....))))))))).-)-----))))....)))))))))...........----...............---...(((((((...))))))). ( -23.10) >DroPse_CAF1 11374 99 + 1 UGUGAAAAAUAGAGCACAUUGAAAUUAA-AACCUCUUAACAUUUGUUUUGUUCA---GUUUUUGAAUUUCAAUU-AU----UGUUUUUUGU--UUUGGUUUUUGUGUGCU---------- ............(((((((.((((((((-(((.....((((.(((....(((((---(...))))))..)))..-..----))))....))--))))))))).)))))))---------- ( -23.30) >DroSec_CAF1 10397 103 + 1 UGCGAA-AAUAGAGCACU-UGAAAACAAAAACUUGUUCAAAU-U-----GUUCACUCGUUUUCGUAUCUCUAUUUGC----UGUUUUUUUU--UUU---UAUUUUGUGCUAGGCACAAAC ((((((-(((.(((((.(-(((..((((....))))))))..-)-----))))....)))))))))...........----..........--...---...(((((((...))))))). ( -20.20) >DroEre_CAF1 11069 103 + 1 UGUGAA-AAUAGAGCACUUUGAAAACAAAAACUUGUUCAAAU-U-----GUUCACUCAUUUUCGUAUCUCGAUUUGC----UGCUUUUU-U--UUU---UAUUUUGUGCUAGGCACAAAC .(..((-....(((((.(((((..((((....))))))))).-)-----))))........(((.....)))))..)----........-.--...---...(((((((...))))))). ( -19.50) >DroYak_CAF1 10413 108 + 1 UGUGAA-AAUAGAGCACUUUGAAAACAAAAACUUGUUCAAAUUU-----GUUCACUCGUUUUCGUAUCUCUAUUUGCAAUAUAUUUUUU-U--UUU---AAUUCCGUGCUAUGCACAAAC ((((..-.....(((((.(((((((.(((((..(((((((((..-----.......((....)).......))))).))))..))))))-)--)))---))....)))))...))))... ( -17.29) >consensus UGUGAA_AAUAGAGCACUUUGAAAACAAAAACUUGUUCAAAU_U_____GUUCACUCGUUUUCGUAUCUCUAUUUGC____UGUUUUUU_U__UUU___UAUUUUGUGCUAGGCACAAAC ............(((((...((((((...............................))))))(((........)))............................))))).......... ( -5.90 = -6.26 + 0.36)


| Location | 16,295,819 – 16,295,925 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -20.72 |
| Consensus MFE | -10.47 |
| Energy contribution | -11.67 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16295819 106 - 22224390 GUUUGUGCCUAGCACAAAAUA---AAAGAAAAAAAAACA----GCAAAUAGAGAUACGAAAACGAGUGAAC-----A-AUUUGAACAAGUUUUUGUUUUCAAAGUGCUCUAUU-UUCGCA .(((((((...)))))))...---...............----((((((((((.(((((((((((..((((-----.-..........))))))))))))...))))))))))-)..)). ( -23.40) >DroPse_CAF1 11374 99 - 1 ----------AGCACACAAAAACCAAA--ACAAAAAACA----AU-AAUUGAAAUUCAAAAAC---UGAACAAAACAAAUGUUAAGAGGUU-UUAAUUUCAAUGUGCUCUAUUUUUCACA ----------((((((...........--..........----..-..((((((((..(((((---(.((((.......))))....))))-))))))))))))))))............ ( -14.11) >DroSec_CAF1 10397 103 - 1 GUUUGUGCCUAGCACAAAAUA---AAA--AAAAAAAACA----GCAAAUAGAGAUACGAAAACGAGUGAAC-----A-AUUUGAACAAGUUUUUGUUUUCA-AGUGCUCUAUU-UUCGCA .(((((((...)))))))...---...--..........----((((((((((.(((((((((((..((((-----.-..........)))))))))))).-.))))))))))-)..)). ( -23.80) >DroEre_CAF1 11069 103 - 1 GUUUGUGCCUAGCACAAAAUA---AAA--A-AAAAAGCA----GCAAAUCGAGAUACGAAAAUGAGUGAAC-----A-AUUUGAACAAGUUUUUGUUUUCAAAGUGCUCUAUU-UUCACA .(((((((...)))))))...---...--(-(((.((.(----(((....((((.((((((((..((....-----.-......))..))))))))))))....)))))).))-)).... ( -20.10) >DroYak_CAF1 10413 108 - 1 GUUUGUGCAUAGCACGGAAUU---AAA--A-AAAAAAUAUAUUGCAAAUAGAGAUACGAAAACGAGUGAAC-----AAAUUUGAACAAGUUUUUGUUUUCAAAGUGCUCUAUU-UUCACA .(((((((...)))))))...---...--.-...........((.((((((((.((((((((((((....)-----((((((....))))))))))))))...))))))))))-).)).. ( -22.20) >consensus GUUUGUGCCUAGCACAAAAUA___AAA__A_AAAAAACA____GCAAAUAGAGAUACGAAAACGAGUGAAC_____A_AUUUGAACAAGUUUUUGUUUUCAAAGUGCUCUAUU_UUCACA .(((((((...))))))).........................(((....((((.((((((((..((.................))..))))))))))))....)))............. (-10.47 = -11.67 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:17 2006