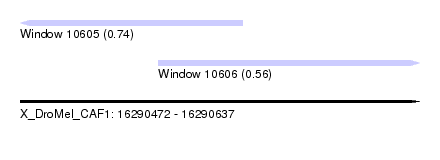
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,290,472 – 16,290,637 |
| Length | 165 |
| Max. P | 0.739556 |

| Location | 16,290,472 – 16,290,564 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -21.46 |
| Energy contribution | -21.42 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16290472 92 - 22224390 AAUUCCUUCA-ACCCGAGCCA----GAGCCGUUGUUGAGCAACUCUAUUUGAUAGCAACUUCCCCGGCUACAAAUUCUUAGUGGUUCAAA--UGGCUAU ..........-.....(((((----(((((((((.....))))...(((((.((((..........))))))))).......)))))...--))))).. ( -21.90) >DroPse_CAF1 5917 91 - 1 --------CAAGUCCGAACAAAGUCCAGCCGUUGUCGAGCAACUCUAUUUGAUAGCAACUUCCCCGGCUACAAAUUCUUAGUGGUUCAGAACUGGCUGU --------.................((((((((.(.((((.(((..(((((.((((..........)))))))))....))).))))).))).))))). ( -22.30) >DroSec_CAF1 5123 92 - 1 AAUUCCUACA-AACCGAGCCA----GAGCCGUUGUUGAGCAACUCUAUUUGAUAGCAACUUCCCCGGCUACAAAUUCUUAGUGGUUCAAA--UGGCUAU ..........-.....(((((----(((((((((.....))))...(((((.((((..........))))))))).......)))))...--))))).. ( -21.90) >DroEre_CAF1 5901 90 - 1 A--UCCCCCA-ACCCGAGCCA----GAGCCGUUGUUGAGCAACUCUAUUUGAUAGCAACUUCCCCGGCUACAAAUUCUUAGUGGUUCAGA--UGGCUAU .--.......-..........----.(((((((..(((((.(((..(((((.((((..........)))))))))....))).)))))))--))))).. ( -21.90) >DroYak_CAF1 5128 91 - 1 A--UCCCCCAAACCCGAGCCA----GAGCCGUUGUUGAGCAACUCUAUUUGAUAGCAACUUCCCCGGCUACAAAUUCUUAGUGGUUCAGA--UGGCUAU .--..................----.(((((((..(((((.(((..(((((.((((..........)))))))))....))).)))))))--))))).. ( -21.90) >consensus A__UCCCCCA_ACCCGAGCCA____GAGCCGUUGUUGAGCAACUCUAUUUGAUAGCAACUUCCCCGGCUACAAAUUCUUAGUGGUUCAGA__UGGCUAU ..........................(((((...((((((.(((..(((((.((((..........)))))))))....))).))))))...))))).. (-21.46 = -21.42 + -0.04)

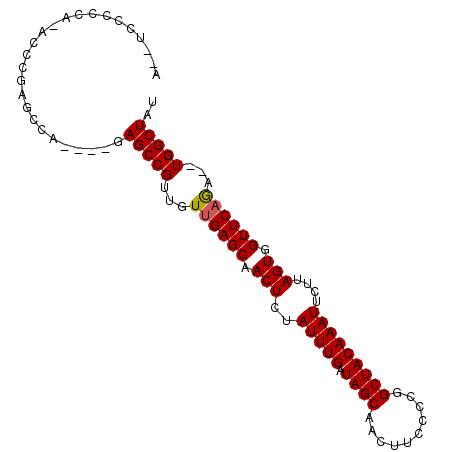
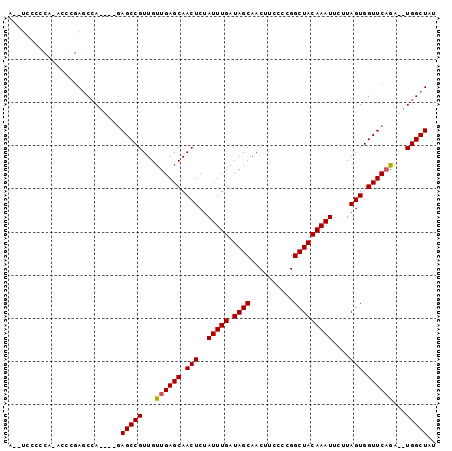
| Location | 16,290,529 – 16,290,637 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 86.10 |
| Mean single sequence MFE | -39.09 |
| Consensus MFE | -27.15 |
| Energy contribution | -29.40 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16290529 108 + 22224390 GCUCAACAACGGCUCUGGCUCGGGU-UGAAGGAAUUUUUGGAGGUCGAGGGGAGCCAACGACACAACGCAGGCUGCCCC---A----AAAAAACGGGCUAAGAAAUCAGGUUGGGC (((((((......(((((((((..(-(...((..((....))..))..((((((((..((......))..)))).))))---.----..))..)))))).)))......))))))) ( -36.60) >DroSec_CAF1 5180 105 + 1 GCUCAACAACGGCUCUGGCUCGGUU-UGUAGGAAUUUUUCGAGGUCGAGGGGUGCCAGCGACACAACGCAGGCUCCCC---------AAAAAACGGGCUUAGAAAUCAGGU-GGGC (((((........(((((((((...-............(((....)))((((.(((.(((......))).))).))))---------......))))).)))).......)-)))) ( -36.76) >DroEre_CAF1 5958 104 + 1 GCUCAACAACGGCUCUGGCUCGGGU-UGGGGGA--UCUAGGAGGUCGAGGGGAGCCAACGACACAACGCAGGCUCCCCC---------AAAAACUGGCUAAGAAAUCAGGGUGGCC .........(.(((((((....(((-..(..((--((.....))))(.((((((((..((......))..)))))))))---------.....)..)))......))))))).).. ( -37.90) >DroYak_CAF1 5185 114 + 1 GCUCAACAACGGCUCUGGCUCGGGUUUGGGGGA--UCUAGGAGGUCGAGGGGAGCCAACGACACAACGCAGGCUCCCCCCCCAAAAAUAAAAACUGGCUAAGAAAUCAGGGUGGGC (((((........((((((.(((.(((((((((--((.....))))..((((((((..((......))..)))))))))))))))........)))))).)))........))))) ( -45.09) >consensus GCUCAACAACGGCUCUGGCUCGGGU_UGGAGGA__UCUAGGAGGUCGAGGGGAGCCAACGACACAACGCAGGCUCCCCC________AAAAAACGGGCUAAGAAAUCAGGGUGGGC (((((((......(((((((((.........((.(((...))).))..((((((((..((......))..))))))))...............)))))).)))......))))))) (-27.15 = -29.40 + 2.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:13 2006