
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,265,109 – 16,265,266 |
| Length | 157 |
| Max. P | 0.917209 |

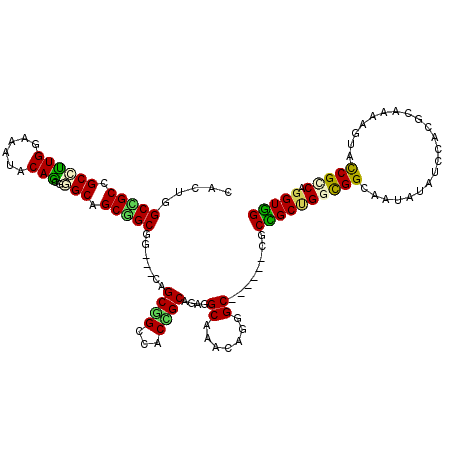
| Location | 16,265,109 – 16,265,226 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -54.68 |
| Consensus MFE | -26.64 |
| Energy contribution | -25.42 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
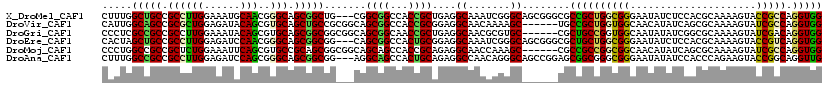
>X_DroMel_CAF1 16265109 117 + 22224390 CUUUGGCUGCCGCCUUGGAAAUGCAACGGGCAGCGGCUG---CGGCGGCCACCGCUGAGGCAAAUCGGGCAGCGGGCGCCGCUGGCGGGAAUAUCUCCACGCAAAAGUACCGCCAGGUGG ...((((((((((..(.(...(((.....))).).)..)---)))))))))(((((..(((........((((((...))))))((((((.....))).))).........))).))))) ( -53.10) >DroVir_CAF1 9572 114 + 1 CAUUGGCAGCCGCGCUGGAGAUACAGCGUGCAGCUGCCGCGGCAGCGGCCACCGCGGAGGCAACAAAAGC------UGCCGCUGGUGGCAACAUAUCAGCGCAAAAGUAUCGCCAGGUGG .........((((.((((.(((((...((((.(((((....))))).((((((((((.(((.......))------).)))).)))))).........))))....))))).)))))))) ( -57.30) >DroGri_CAF1 9753 114 + 1 CCCUCGCCGCCGCCUUGGAAAUACAGCGUGCAGCGGCGGCGGCAGCGGCAACCGCUGAGGCAACGCGUGC------CGCUGCCGGUGGCAAUAUAUCGGCGCAAAAGUAUCGACAGGUGG ....(((((((((((((......))).(.((((((((((((.((((((...))))))......))).)))------)))))))))))))......(((..((....))..)))..)))). ( -54.80) >DroEre_CAF1 2292 117 + 1 CACUAGCUGCCGCCUUGGAGAUCCAACGGGCAGCGGCGG---CAGCGGCCACUGCGGAGGCAAAUCGGGCAGCGGGCGCUGCUGGCGGGAAUAUCUCCACGCAAAAGUACCGUCAGGUGG ((((.((((((.(.((((....)))).)))))))(((((---(((((.((.((((.((......))..)))).)).))))))))((((((.....))).))).......))....)))). ( -57.90) >DroMoj_CAF1 6320 114 + 1 CCCUGGCCGCCGCUCUGGAAAUUCAGCGUGCCGCAGCGGCGGCAGCAGCCACCGCAGAGGCAACCAAAGC------CGCCGCCGGCGGCAACAUAUCAGCGCAAAAGUAUCGCCAGGUGG .((((((..((.....)).......(((((((...((((.(((....))).))))...))).......((------((((...)))))).........)))).........))))))... ( -49.40) >DroAna_CAF1 8481 117 + 1 CUUUGGCCGCCGCCUUGGAGAUCCAGCGGGCAGCGGCGG---AGGCAGCCACUGCAGAGGCCAACAGGGCAGCCGGAGCGGCGGGCGGGAAUAUAUCCACCCAGAAGUACCGGCAGGUUG ((((.(((((.(((((((....)))..)))).))))).)---)))(((((.........(((.....))).(((((.((..((((.(((......))).))).)..)).))))).))))) ( -55.60) >consensus CACUGGCCGCCGCCUUGGAAAUACAGCGGGCAGCGGCGG___CAGCGGCCACCGCAGAGGCAAACAGGGC______CGCCGCUGGCGGCAAUAUAUCCACGCAAAAGUACCGCCAGGUGG .....(((((.((((((......)))..))).))))).......((((...))))....((.......))........((((((((((.....................))))).))))) (-26.64 = -25.42 + -1.22)

| Location | 16,265,109 – 16,265,226 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.09 |
| Mean single sequence MFE | -55.57 |
| Consensus MFE | -23.67 |
| Energy contribution | -23.40 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
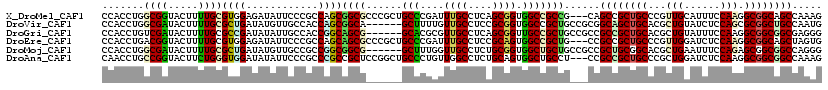
>X_DroMel_CAF1 16265109 117 - 22224390 CCACCUGGCGGUACUUUUGCGUGGAGAUAUUCCCGCCAGCGGCGCCCGCUGCCCGAUUUGCCUCAGCGGUGGCCGCCG---CAGCCGCUGCCCGUUGCAUUUCCAAGGCGGCAGCCAAAG .....(((((((......(((.((((...)))))))..((((((.(((((((..((......)).))))))).)))))---).)))((((((..(((......))))))))).))))... ( -53.10) >DroVir_CAF1 9572 114 - 1 CCACCUGGCGAUACUUUUGCGCUGAUAUGUUGCCACCAGCGGCA------GCUUUUGUUGCCUCCGCGGUGGCCGCUGCCGCGGCAGCUGCACGCUGUAUCUCCAGCGCGGCUGCCAAUG .....((((((((..((......))..))))))))...((((((------((....((..((.....))..)).))))))))(((((((((..((((......))))))))))))).... ( -57.10) >DroGri_CAF1 9753 114 - 1 CCACCUGUCGAUACUUUUGCGCCGAUAUAUUGCCACCGGCAGCG------GCACGCGUUGCCUCAGCGGUUGCCGCUGCCGCCGCCGCUGCACGCUGUAUUUCCAAGGCGGCGGCGAGGG ...(((((((............)))))..(((((...(((((((------((((.(((((...)))))).))))))))))((((((((.....))((......)).))))))))))))). ( -52.70) >DroEre_CAF1 2292 117 - 1 CCACCUGACGGUACUUUUGCGUGGAGAUAUUCCCGCCAGCAGCGCCCGCUGCCCGAUUUGCCUCCGCAGUGGCCGCUG---CCGCCGCUGCCCGUUGGAUCUCCAAGGCGGCAGCUAGUG .(((.....(((......(((.((((...)))))))..((((((.(((((((..((......)).))))))).)))))---).)))(((((((.((((....)))).).))))))..))) ( -53.60) >DroMoj_CAF1 6320 114 - 1 CCACCUGGCGAUACUUUUGCGCUGAUAUGUUGCCGCCGGCGGCG------GCUUUGGUUGCCUCUGCGGUGGCUGCUGCCGCCGCUGCGGCACGCUGAAUUUCCAGAGCGGCGGCCAGGG ...((((((((.....)).(((((......((((((.(((((((------((..((((..((.....))..))))..)))))))))))))))..(((......)))..))))))))))). ( -62.00) >DroAna_CAF1 8481 117 - 1 CAACCUGCCGGUACUUCUGGGUGGAUAUAUUCCCGCCCGCCGCUCCGGCUGCCCUGUUGGCCUCUGCAGUGGCUGCCU---CCGCCGCUGCCCGCUGGAUCUCCAAGGCGGCGGCCAAAG ......(((((......(((((((........))))))).....))))).......((((((...((((((((.....---..))))))))(((((((....))..))))).)))))).. ( -54.90) >consensus CCACCUGGCGAUACUUUUGCGCGGAUAUAUUCCCGCCAGCGGCG______GCCCGAGUUGCCUCAGCGGUGGCCGCUG___CCGCCGCUGCACGCUGGAUCUCCAAGGCGGCGGCCAAAG .......((((.....))))((((............))))((((......(((....((((....)))).)))))))......((((((((...(((......))).))))))))..... (-23.67 = -23.40 + -0.27)
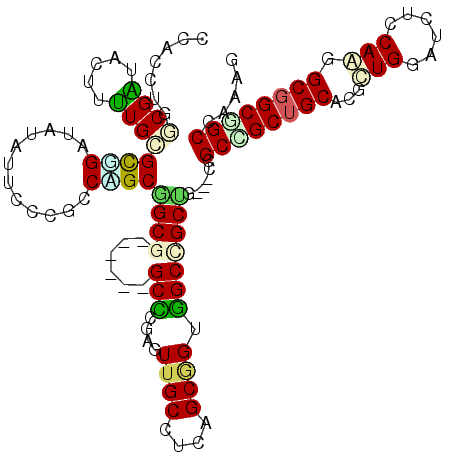
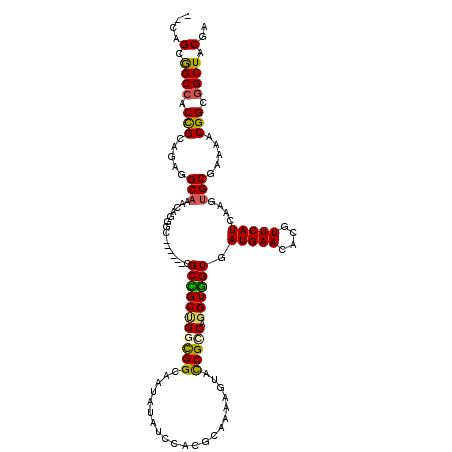
| Location | 16,265,148 – 16,265,266 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -45.60 |
| Consensus MFE | -26.21 |
| Energy contribution | -25.43 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16265148 118 + 22224390 --CGGCGGCCACCGCUGAGGCAAAUCGGGCAGCGGGCGCCGCUGGCGGGAAUAUCUCCACGCAAAAGUACCGCCAGGUGGUGAUGAACGCGUUCAUUAAGUGCGAGGGCGGCGGCUACGA --(((((((..((((((.((....))...))))))..)))))))((((((.....))).)))....((((((((......(((((((....)))))))...((....))))))).))).. ( -52.90) >DroVir_CAF1 9612 114 + 1 GGCAGCGGCCACCGCGGAGGCAACAAAAGC------UGCCGCUGGUGGCAACAUAUCAGCGCAAAAGUAUCGCCAGGUGGUCAUGAACACGUUCAUCAAGUGCGAGAACGGCGGCUUCGA (((.(((((((((((((.((((........------))))(((((((....)))..)))).........))))..))))))).......(((((.((......))))))))).))).... ( -43.80) >DroGri_CAF1 9793 114 + 1 GGCAGCGGCAACCGCUGAGGCAACGCGUGC------CGCUGCCGGUGGCAAUAUAUCGGCGCAAAAGUAUCGACAGGUGGUGAUGAACACGUUCAUCAAAUGCGAAAACGGUGGCUUCGA ((((((((((..(((...(....)))))))------))))))).(.(((....(((((.((((....((((....)))).(((((((....)))))))..))))....)))))))).).. ( -45.30) >DroEre_CAF1 2331 118 + 1 --CAGCGGCCACUGCGGAGGCAAAUCGGGCAGCGGGCGCUGCUGGCGGGAAUAUCUCCACGCAAAAGUACCGUCAGGUGGUGAUGAACGCGUUCAUUAAGUGCGAAGGCGGCGGCUACGA --(((((((..((((....((.......)).))))..)))))))((((((.....))).)))....((((((((......(((((((....)))))))...((....))))))).))).. ( -47.10) >DroMoj_CAF1 6360 114 + 1 GGCAGCAGCCACCGCAGAGGCAACCAAAGC------CGCCGCCGGCGGCAACAUAUCAGCGCAAAAGUAUCGCCAGGUGGUAAUGAAUACAUUCAUCAAAUGCGAAAACGGCGGCUUCGA (((....)))......(((((..((...((------((((...))))))..........((((....((((((...))))))(((((....)))))....)))).....))..))))).. ( -37.30) >DroAna_CAF1 8520 118 + 1 --AGGCAGCCACUGCAGAGGCCAACAGGGCAGCCGGAGCGGCGGGCGGGAAUAUAUCCACCCAGAAGUACCGGCAGGUUGUGAUGAACGCGUUCAUAAAGUGCGAGGGCGGUGGCUACGA --..(.(((((((((....(((..((.(((.(((((.((..((((.(((......))).))).)..)).)))))..))).))(((((....)))))...).))....))))))))).).. ( -47.20) >consensus __CAGCGGCCACCGCAGAGGCAAACAGGGC______CGCCGCUGGCGGCAAUAUAUCCACGCAAAAGUACCGCCAGGUGGUGAUGAACACGUUCAUCAAGUGCGAAAACGGCGGCUACGA ....(.((((.(((.....(((...............(((((((((((.....................))))).)))))).(((((....)))))....))).....))).)))).).. (-26.21 = -25.43 + -0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:09 2006