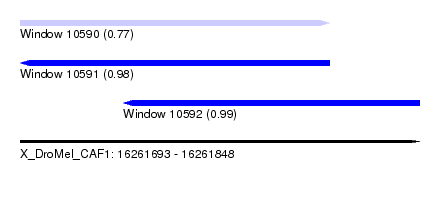
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,261,693 – 16,261,848 |
| Length | 155 |
| Max. P | 0.994958 |

| Location | 16,261,693 – 16,261,813 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.32 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
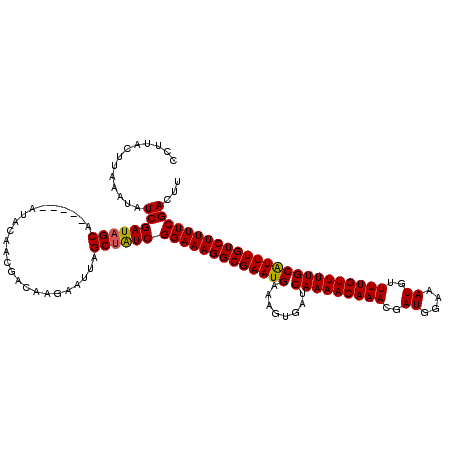
>X_DroMel_CAF1 16261693 120 + 22224390 AACACACCAACACAAACAAACGCAUUUGCACACGUAGCUGAAGUCGAAAAGACAAAUGCAAAACAAAACAUUUCCAUCGUUUGUUUUGAUCACUUUCAUUCGCCUUUCCGACAGCUAAUU .....................((....)).....(((((...((((.((((.(.(((((((((((((............)))))))))........)))).).)))).)))))))))... ( -19.50) >DroSec_CAF1 9990 120 + 1 AACACACCAACACAAACAAACGCAUGUGCACACAUAGCUGAAGUCGAAAAGACAAAUGCAAAACAAAACAUUUCCAUCUUUUGUUUUGAUCACUUUCAUUCGCCUUUCCGACAGCUAAUU .....................((....)).....(((((...((((.((((.(.((((((((((((((.((....)).))))))))))........)))).).)))).)))))))))... ( -20.80) >DroEre_CAF1 10100 120 + 1 AACACACCAACACAAACAAACGCAUGUGCACGCAUAGCUGAAGUCGAAAAGACAAACGCAAAACAAAACAUUUCCAUCGUUUGUUUUGAUCACUUUCAUUCGCCUUUCCGAUUGCUAAUU .....................((....))..(((.....((((.((((((((((((((...................))))))))))((......)).)))).)))).....)))..... ( -18.11) >DroYak_CAF1 9995 120 + 1 AACACACCAACACAAACAAACGCAUGUGCACACAUAGCUAAAGUCGAAAAGACAAAUGCAAAACAAAACAUUUCCAUCGUUUGUUUUGAUCACUUUCAUUCGCCUUUCCGAUAGCUAAUU .....................((....)).....(((((...((((.((((.(.(((((((((((((............)))))))))........)))).).)))).)))))))))... ( -18.30) >consensus AACACACCAACACAAACAAACGCAUGUGCACACAUAGCUGAAGUCGAAAAGACAAAUGCAAAACAAAACAUUUCCAUCGUUUGUUUUGAUCACUUUCAUUCGCCUUUCCGACAGCUAAUU .....................((....)).....((((((..(((.....))).....(((((((((............))))))))).......................))))))... (-17.51 = -17.32 + -0.19)



| Location | 16,261,693 – 16,261,813 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -34.61 |
| Energy contribution | -34.92 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16261693 120 - 22224390 AAUUAGCUGUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAACGAUGGAAAUGUUUUGUUUUGCAUUUGUCUUUUCGACUUCAGCUACGUGUGCAAAUGCGUUUGUUUGUGUUGGUGUGUU ...(((((((((((((((((((((........(((((((((..((....))..)))))))))))))))))))))))))...))))).(..(((.((((((......)))))).)))..). ( -36.80) >DroSec_CAF1 9990 120 - 1 AAUUAGCUGUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAAAGAUGGAAAUGUUUUGUUUUGCAUUUGUCUUUUCGACUUCAGCUAUGUGUGCACAUGCGUUUGUUUGUGUUGGUGUGUU ...(((((((((((((((((((((........((((((((((.((....)).))))))))))))))))))))))))))...))))).....(((((((((......)))....)))))). ( -41.00) >DroEre_CAF1 10100 120 - 1 AAUUAGCAAUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAACGAUGGAAAUGUUUUGUUUUGCGUUUGUCUUUUCGACUUCAGCUAUGCGUGCACAUGCGUUUGUUUGUGUUGGUGUGUU .(((((((.(((((((((((((((........(((((((((..((....))..))))))))))))))))))))))))....(((.((((((....))))))..)))..)))))))..... ( -34.50) >DroYak_CAF1 9995 120 - 1 AAUUAGCUAUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAACGAUGGAAAUGUUUUGUUUUGCAUUUGUCUUUUCGACUUUAGCUAUGUGUGCACAUGCGUUUGUUUGUGUUGGUGUGUU ...(((((((((((((((((((((........(((((((((..((....))..))))))))))))))))))))))))...)))))).....(((((((((......)))....)))))). ( -35.90) >consensus AAUUAGCUAUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAACGAUGGAAAUGUUUUGUUUUGCAUUUGUCUUUUCGACUUCAGCUAUGUGUGCACAUGCGUUUGUUUGUGUUGGUGUGUU ...(((((((((((((((((((((........(((((((((..((....))..))))))))))))))))))))))))...))))))...(..((((((((......)))))..)))..). (-34.61 = -34.92 + 0.31)


| Location | 16,261,733 – 16,261,848 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -27.47 |
| Energy contribution | -27.29 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16261733 115 - 22224390 UCAUACUUGAAUGUCGAUAGCA-----AUACAACGGCAAGAAUUAGCUGUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAACGAUGGAAAUGUUUUGUUUUGCAUUUGUCUUUUCGACUU (((....)))..((((((((((-----((.(........).))).))))))(((((((((((((........(((((((((..((....))..))))))))))))))))))))))))).. ( -32.60) >DroSec_CAF1 10030 115 - 1 UUCAACUUGAAUAUCGAUAGCA-----AUACAACGACUAGAAUUAGCUGUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAAAGAUGGAAAUGUUUUGUUUUGCAUUUGUCUUUUCGACUU .............(((((((((-----((.(........).))).))))))(((((((((((((........((((((((((.((....)).)))))))))))))))))))))))))... ( -32.90) >DroEre_CAF1 10140 120 - 1 CCUUACUUAAAUAUCGAUAGCACAGCACUACAGCGACAAGAAUUAGCAAUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAACGAUGGAAAUGUUUUGUUUUGCGUUUGUCUUUUCGACUU ........................((......))...............(((((((((((((((........(((((((((..((....))..))))))))))))))))))))))))... ( -25.80) >DroYak_CAF1 10035 112 - 1 CCUCACUUAAAUAUCGAUAGCA-----AUACA---ACAAUAAUUAGCUAUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAACGAUGGAAAUGUUUUGUUUUGCAUUUGUCUUUUCGACUU .............((((((((.-----.....---..........))))))(((((((((((((........(((((((((..((....))..))))))))))))))))))))))))... ( -28.59) >consensus CCUUACUUAAAUAUCGAUAGCA_____AUACAACGACAAGAAUUAGCUAUCGGAAAGGCGAAUGAAAGUGAUCAAAACAAACGAUGGAAAUGUUUUGUUUUGCAUUUGUCUUUUCGACUU .............((((((((........................))))))(((((((((((((........(((((((((..((....))..))))))))))))))))))))))))... (-27.47 = -27.29 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:15:00 2006