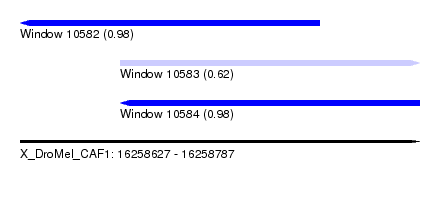
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,258,627 – 16,258,787 |
| Length | 160 |
| Max. P | 0.982500 |

| Location | 16,258,627 – 16,258,747 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.66 |
| Mean single sequence MFE | -40.36 |
| Consensus MFE | -38.42 |
| Energy contribution | -38.46 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16258627 120 - 22224390 AUGCGCGCGCGCAAAGAGAGAGCGCCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUACGCAGCAGGCAGCUACAGCGCAAAAUUAUAUGUAAUUGGCC .(((((..((((.........))(((((.((.((((((.....)).)))))).))..(((.....((((....))))..)))....))).))....)))))................... ( -37.40) >DroSec_CAF1 6941 119 - 1 AUGCGCGCGCGCAAAGAGAGAGCGGCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUACGCAGCAGGCAGC-ACAGCGCAAAAUUAUAUGUAAUUGGCC .((((((.((((.....(.(((.((((((((.((.....))..))))))))))))..(((.....((((....))))..))).....)).))-.).)))))................... ( -41.00) >DroSim_CAF1 7082 119 - 1 AUGCGCGCGCGCAAAGAGAGAGCGGCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUACGCAGCAGGCAGC-ACAGCGCAAAAUUAUAUGUAAUUGGCC .((((((.((((.....(.(((.((((((((.((.....))..))))))))))))..(((.....((((....))))..))).....)).))-.).)))))................... ( -41.00) >DroEre_CAF1 7067 119 - 1 AUGCGCGCGCGCAAAGAGAGAGCGGCGGUGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGGAAUUCCUUGAAAAAGGUACGCAGCAGGCAGC-ACAGCGCAAAAUUAUAUGUAAUUGGCC .((((((.((((...(.(((.((((((((...((.....)).)))))))).))))..(((.....((((....))))..))).....)).))-.).)))))................... ( -41.40) >DroYak_CAF1 6918 119 - 1 AUGCGCGCGCGCAAAGAGAGAGCGGCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUACGCAGCAGGCAGC-ACAGCGCAAAAUUAUAUGUAAUUGGCC .((((((.((((.....(.(((.((((((((.((.....))..))))))))))))..(((.....((((....))))..))).....)).))-.).)))))................... ( -41.00) >consensus AUGCGCGCGCGCAAAGAGAGAGCGGCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUACGCAGCAGGCAGC_ACAGCGCAAAAUUAUAUGUAAUUGGCC .(((((..((((.....(.(((.((((((((.((.....))..))))))))))))..(((.....((((....))))..))).....)).))....)))))................... (-38.42 = -38.46 + 0.04)

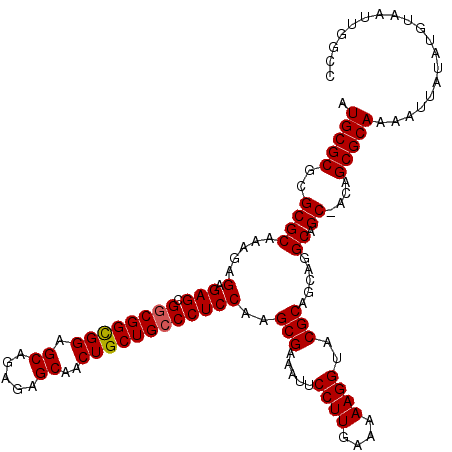
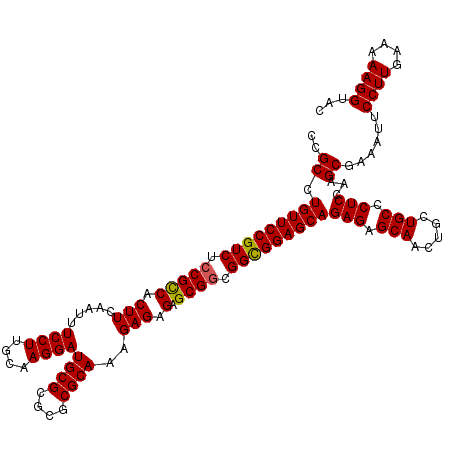
| Location | 16,258,667 – 16,258,787 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -47.08 |
| Consensus MFE | -44.86 |
| Energy contribution | -45.50 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16258667 120 + 22224390 GUACCUUUUUCAAGGAAUUUCGCUUGGAGGGCAGCAGUUGCUCUCUGCUCCGCCGGCGCUCUCUCUUUGCGCGCGCGCAUCCUUGCAAGGAAAUUGAAGUGACGGAGACGGAACAGGCGG ...((((....)))).....((((((..((((((...))))))((((((((((((((((.........)))).)).)).((((....))))............)))).)))).)))))). ( -42.30) >DroSec_CAF1 6980 120 + 1 GUACCUUUUUCAAGGAAUUUCGCUUGGAGGGCAGCAGUUGCUCUCUGCUCCGCCGCCGCUCUCUCUUUGCGCGCGCGCAUCCUUGCAAGGAAAUUGAAGUGGCGGAGACGGAACAGGCUG ...((((....))))......(((((((((((((...))))))))...((((((((((((.((....((((....))))((((....))))....))))))))))...)))).))))).. ( -46.40) >DroSim_CAF1 7121 120 + 1 GUACCUUUUUCAAGGAAUUUCGCUUGGAGGGCAGCAGUUGCUCUCUGCUCCGCCGCCGCUCUCUCUUUGCGCGCGCGCAUCCUUGCAAGGAAAUUGAAGUGGCGGAGACGGAACAGGCUG ...((((....))))......(((((((((((((...))))))))...((((((((((((.((....((((....))))((((....))))....))))))))))...)))).))))).. ( -46.40) >DroEre_CAF1 7106 120 + 1 GUACCUUUUUCAAGGAAUUCCGCUUGGAGGGCAGCAGUUGCUCUCUGCUCCACCGCCGCUCUCUCUUUGCGCGCGCGCAUCCUUGCAAGGAAAUGGAAGUGGCGGAGACGGAACAGGCGG ...((((....))))....(((((((((((((((...))))))))...(((.((((((((..(....((((....))))((((....))))...)..))))))))....))).))))))) ( -49.80) >DroYak_CAF1 6957 120 + 1 GUACCUUUUUCAAGGAAUUUCGCUUGGAGGGCAGCAGUUGCUCUCUGCUCCGCCGCCGCUCUCUCUUUGCGCGCGCGCAUCCUUGCGAGGAAAUGGAAGUGGCGGAGACGGAACAGGCGG ...((((....)))).....((((((((((((((...))))))))...((((((((((((..(....((((....))))((((....))))...)..))))))))...)))).)))))). ( -50.50) >consensus GUACCUUUUUCAAGGAAUUUCGCUUGGAGGGCAGCAGUUGCUCUCUGCUCCGCCGCCGCUCUCUCUUUGCGCGCGCGCAUCCUUGCAAGGAAAUUGAAGUGGCGGAGACGGAACAGGCGG ...((((....))))....(((((((((((((((...))))))))...((((((((((((.((....((((....))))((((....))))....))))))))))...)))).))))))) (-44.86 = -45.50 + 0.64)



| Location | 16,258,667 – 16,258,787 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -44.38 |
| Consensus MFE | -42.08 |
| Energy contribution | -41.96 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16258667 120 - 22224390 CCGCCUGUUCCGUCUCCGUCACUUCAAUUUCCUUGCAAGGAUGCGCGCGCGCAAAGAGAGAGCGCCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUAC .(((.(((((((((..(((...(((....((((....))))((((....))))..)))...)))..)))))))))(((.(((.....))).)))...))).....((((....))))... ( -39.90) >DroSec_CAF1 6980 120 - 1 CAGCCUGUUCCGUCUCCGCCACUUCAAUUUCCUUGCAAGGAUGCGCGCGCGCAAAGAGAGAGCGGCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUAC ..((.(((((((((.(((((.(((.....((((....))))((((....))))..))).).)))).)))))))))(((.(((.....))).)))...))......((((....))))... ( -44.30) >DroSim_CAF1 7121 120 - 1 CAGCCUGUUCCGUCUCCGCCACUUCAAUUUCCUUGCAAGGAUGCGCGCGCGCAAAGAGAGAGCGGCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUAC ..((.(((((((((.(((((.(((.....((((....))))((((....))))..))).).)))).)))))))))(((.(((.....))).)))...))......((((....))))... ( -44.30) >DroEre_CAF1 7106 120 - 1 CCGCCUGUUCCGUCUCCGCCACUUCCAUUUCCUUGCAAGGAUGCGCGCGCGCAAAGAGAGAGCGGCGGUGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGGAAUUCCUUGAAAAAGGUAC ((((.(((((((...(((((.((..(...((((....))))((((....))))....)..)).))))))))))))(((.(((.....))).)))...))))....((((....))))... ( -46.80) >DroYak_CAF1 6957 120 - 1 CCGCCUGUUCCGUCUCCGCCACUUCCAUUUCCUCGCAAGGAUGCGCGCGCGCAAAGAGAGAGCGGCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUAC .(((.(((((((((.((((..((..(...((((....))))((((....))))..)..)).)))).)))))))))(((.(((.....))).)))...))).....((((....))))... ( -46.60) >consensus CCGCCUGUUCCGUCUCCGCCACUUCAAUUUCCUUGCAAGGAUGCGCGCGCGCAAAGAGAGAGCGGCGGCGGAGCAGAGAGCAACUGCUGCCCUCCAAGCGAAAUUCCUUGAAAAAGGUAC ..((.(((((((((.(((((.(((.....((((....))))((((....))))..))).).)))).)))))))))(((.(((.....))).)))...))......((((....))))... (-42.08 = -41.96 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:53 2006