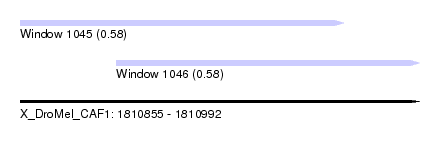
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,810,855 – 1,810,992 |
| Length | 137 |
| Max. P | 0.582671 |

| Location | 1,810,855 – 1,810,966 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 92.10 |
| Mean single sequence MFE | -42.18 |
| Consensus MFE | -36.20 |
| Energy contribution | -35.76 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
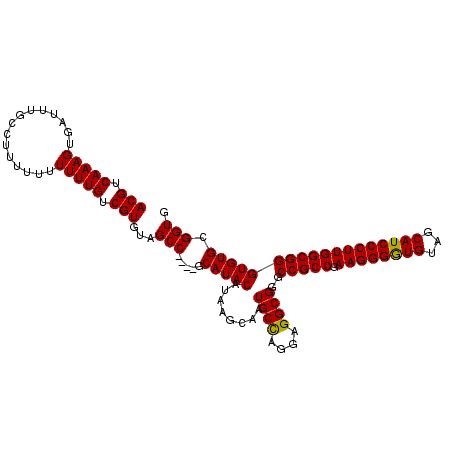
>X_DroMel_CAF1 1810855 111 + 22224390 ACGCCAAAGUGAUUUGACCUUUUUUUUUGUCGUGUAGCCGCCCGCAUACAUAAGCAAUGCCAGGAGGCGGGCCGCCGAAGGGAUGUAGCAUCCCUUGGCGGGUGUGAGGUG ....((((....))))(((((...............(((((((((........))...(((....)))))))((((.((((((((...)))))))))))))))..))))). ( -41.63) >DroSec_CAF1 19238 106 + 1 ACGUCAAAGUGAUUUGCCUUUU-UUUUUGUCGUGUAGCC----ACAUACAUAAGCAAUGCUAGGAGGCGGGCCGCCGAAGGGGUGUAGCAUCCCUUGGCGGGUGUGCGGUG .(((((......((((((((((-...((((.(((((...----...)))))..))))....))))))))))(((((.((((((((...))))))))))))).)).)))... ( -39.10) >DroSim_CAF1 21313 107 + 1 ACGUCAAAGUGAUUUGCCUUUUUUUUUUGUCGUGUAGCC----GCAUACAUAAGCAAUGCCAGGGGGCGGGCCGCCGAAGGGGUGUAGCAUCCCUUGGCGGGUGUGCGGUG (((.(((((.((.........)).))))).)))...(((----((((((........((((....))))..(((((.((((((((...)))))))))))))))))))))). ( -45.80) >consensus ACGUCAAAGUGAUUUGCCUUUUUUUUUUGUCGUGUAGCC____GCAUACAUAAGCAAUGCCAGGAGGCGGGCCGCCGAAGGGGUGUAGCAUCCCUUGGCGGGUGUGCGGUG (((.(((((...............))))).)))...(((.....(((((........((((....))))..(((((.((((((((...)))))))))))))))))).))). (-36.20 = -35.76 + -0.44)

| Location | 1,810,888 – 1,810,992 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 90.91 |
| Mean single sequence MFE | -47.13 |
| Consensus MFE | -39.28 |
| Energy contribution | -38.40 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
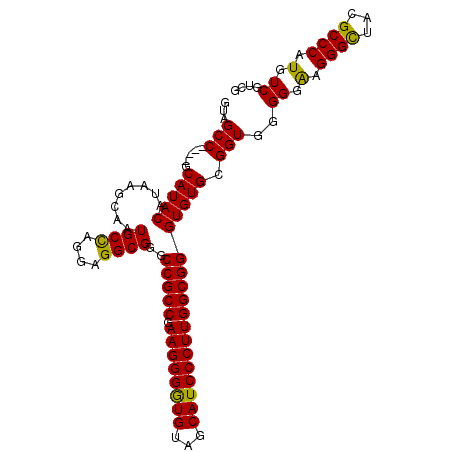
>X_DroMel_CAF1 1810888 104 + 22224390 GUAGCCGCCCGCAUACAUAAGCAAUGCCAGGAGGCGGGCCGCCGAAGGGAUGUAGCAUCCCUUGGCGGGUGUGAGGUGCGGGGAGGGUUCCGCCCAUGUCGUUG ....((((((.(((((........((((....))))..(((((.((((((((...)))))))))))))))))).)).))))...(((.....)))......... ( -48.70) >DroSec_CAF1 19270 100 + 1 GUAGCC----ACAUACAUAAGCAAUGCUAGGAGGCGGGCCGCCGAAGGGGUGUAGCAUCCCUUGGCGGGUGUGCGGUGGGGGAAGGGCUACGCCCAUGUCGUCG ...(((----.(((((........((((....))))..(((((.((((((((...)))))))))))))))))).))).((.((.((((...))))...)).)). ( -42.60) >DroSim_CAF1 21346 100 + 1 GUAGCC----GCAUACAUAAGCAAUGCCAGGGGGCGGGCCGCCGAAGGGGUGUAGCAUCCCUUGGCGGGUGUGCGGUGGGGGAAGGGCUACGCCCAUGUCGUCG ...(((----((((((........((((....))))..(((((.((((((((...)))))))))))))))))))))).((.((.((((...))))...)).)). ( -50.10) >consensus GUAGCC____GCAUACAUAAGCAAUGCCAGGAGGCGGGCCGCCGAAGGGGUGUAGCAUCCCUUGGCGGGUGUGCGGUGGGGGAAGGGCUACGCCCAUGUCGUCG ...(((.....(((((........((((....))))..(((((.((((((((...)))))))))))))))))).)))..((.(.((((...)))).).)).... (-39.28 = -38.40 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:15 2006