
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,258,078 – 16,258,265 |
| Length | 187 |
| Max. P | 0.994577 |

| Location | 16,258,078 – 16,258,185 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 86.99 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -25.49 |
| Energy contribution | -25.93 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16258078 107 + 22224390 -----UUUUGGGUUUCCAAUGUUUCCACUGCCAAGUCACGAUGCAGUGGGAGUGGAAGAGAAUGGGUGUAGGAGAAAGAGGAGACGGAGGAGUAGAAUUGGAGCUC--CAUUCU -----...(((..(((((((.((((((((.(((.((......))..))).)))))))).....................(....)...........)))))))..)--)).... ( -28.80) >DroSec_CAF1 6405 100 + 1 -----UUUUGGGUUUCCAAUGUUUCCACUGCCAAGUCACGAUGCAGUGGAAGUGGAAGAGAAUGGGUGUAGG---------AGACGGAGGAGUAGAAUUGGAGCUCCCCAUUCU -----.......((((((...((((((((((...........))))))))))))))))(((((((.....(.---------...)...(((((.........)))))))))))) ( -34.80) >DroSim_CAF1 6303 109 + 1 -----UUUUGGGUUUCCAAUGUUUCCACUGCCAAGUCACGAUGCAGUGGGAGUGGAAGAGAAUGGGUGUAGGAGAAAGAGGAGACGGAGGAGUAGAAUUGGAGCUCCCCAUUCU -----.......((((((...((((((((((...........))))))))))))))))(((((((..............(....)...(((((.........)))))))))))) ( -34.30) >DroEre_CAF1 6492 103 + 1 CCGCUUUUUGGGUUUCCAAUGUUUCCACUGCCAAGUCACGAUGCAGUGGGAAUGGAAGAGAAUGGGUAU---------AGGAGGAGGAGGAGCAGAAGUGGAGCUC--CAUUCU ((.(((((((..((((((...((((((((((...........))))))))))))))))..........)---------)))))).)).(((((.........))))--)..... ( -32.10) >consensus _____UUUUGGGUUUCCAAUGUUUCCACUGCCAAGUCACGAUGCAGUGGGAGUGGAAGAGAAUGGGUGUAGG______AGGAGACGGAGGAGUAGAAUUGGAGCUC__CAUUCU ............((((((...((((((((((...........))))))))))))))))((((((((.......................((((.........)))))))))))) (-25.49 = -25.93 + 0.44)

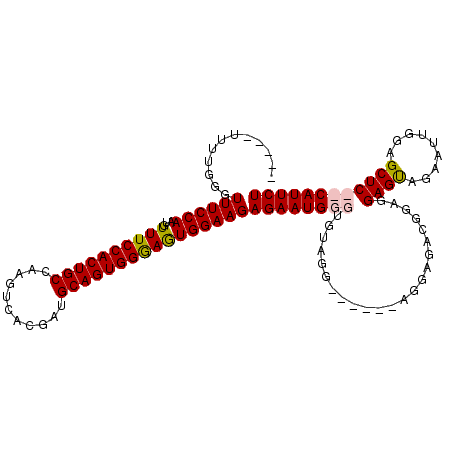

| Location | 16,258,078 – 16,258,185 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.99 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -18.49 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16258078 107 - 22224390 AGAAUG--GAGCUCCAAUUCUACUCCUCCGUCUCCUCUUUCUCCUACACCCAUUCUCUUCCACUCCCACUGCAUCGUGACUUGGCAGUGGAAACAUUGGAAACCCAAAA----- ((((((--(........................................))))))).(((((((.(((...(.....)...))).)))))))...((((....))))..----- ( -20.10) >DroSec_CAF1 6405 100 - 1 AGAAUGGGGAGCUCCAAUUCUACUCCUCCGUCU---------CCUACACCCAUUCUCUUCCACUUCCACUGCAUCGUGACUUGGCAGUGGAAACAUUGGAAACCCAAAA----- (((((((((((...........))))...((..---------...))..))))))).(((((.(((((((((...........)))))))))....)))))........----- ( -28.30) >DroSim_CAF1 6303 109 - 1 AGAAUGGGGAGCUCCAAUUCUACUCCUCCGUCUCCUCUUUCUCCUACACCCAUUCUCUUCCACUCCCACUGCAUCGUGACUUGGCAGUGGAAACAUUGGAAACCCAAAA----- (((((((((((...........))))...(......)............))))))).(((((((.(((...(.....)...))).)))))))...((((....))))..----- ( -24.70) >DroEre_CAF1 6492 103 - 1 AGAAUG--GAGCUCCACUUCUGCUCCUCCUCCUCCU---------AUACCCAUUCUCUUCCAUUCCCACUGCAUCGUGACUUGGCAGUGGAAACAUUGGAAACCCAAAAAGCGG .....(--((((.........)))))..........---------............(((((...(((((((...........)))))))......)))))............. ( -23.20) >consensus AGAAUG__GAGCUCCAAUUCUACUCCUCCGUCUCCU______CCUACACCCAUUCUCUUCCACUCCCACUGCAUCGUGACUUGGCAGUGGAAACAUUGGAAACCCAAAA_____ (((((((((((...........))).......................)))))))).(((((((.(((...(.....)...))).)))))))...((((....))))....... (-18.49 = -19.30 + 0.81)

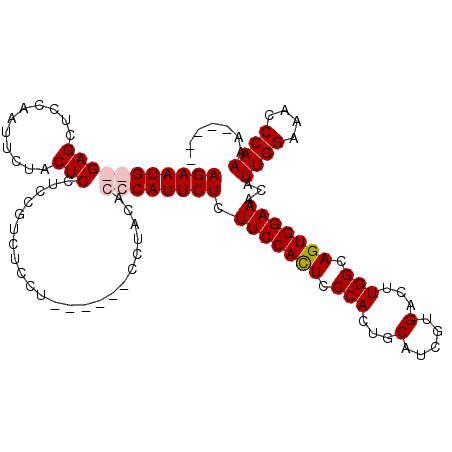
| Location | 16,258,147 – 16,258,265 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -25.74 |
| Energy contribution | -27.80 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.79 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16258147 118 + 22224390 AAAGAGGAGACGGAGGAGUAGAAUUGGAGCUC--CAUUCUGGCUGAAAUCCAAUCACGACCACAAUCUUUAGCUCAUAAUCAAACUUGAACUCUACACUUUUGCUACUCCUCCUUCUUCG .....(((((.((((((((((.....(((((.--.(((.(((((((.......))).).))).)))....)))))....(((....)))..............)))))))))).))))). ( -35.60) >DroSec_CAF1 6472 113 + 1 -------AGACGGAGGAGUAGAAUUGGAGCUCCCCAUUCUGGCUGAAAUCCAAUCACGACCACAAUCUUUAGCUCAUAAUCAAACUUGAACUCUACACUUUUGCUACUCCUCCUCCUUCG -------.((.((((((((((.....(((((....(((.(((((((.......))).).))).)))....)))))....(((....)))..............))))))))))))..... ( -29.90) >DroSim_CAF1 6372 117 + 1 AAAGAGGAGACGGAGGAGUAGAAUUGGAGCUCCCCAUUCUGGCUGAAAUCCAAUCACGACCACAAUCUUUAGCUCAUAAUCAAACUUGAACUCUACACUUGA---AUUCCACCACCAUUG .....((((...(((..(((((....(((((....(((.(((((((.......))).).))).)))....)))))....(((....)))..))))).)))..---.)))).......... ( -21.50) >DroEre_CAF1 6561 114 + 1 ----AGGAGGAGGAGGAGCAGAAGUGGAGCUC--CAUUCUGGCUGAAAUCCAAUCACGACCACAAUCUUUAGCUCAUAAUCAAACUUGAACUCUACACUUUUGCUACUCCUCCUCCAUCC ----.((((((((((.(((((((((((((((.--.(((.(((((((.......))).).))).)))....)))))....(((....)))......)))))))))).)))))))))).... ( -47.80) >consensus ____AGGAGACGGAGGAGUAGAAUUGGAGCUC__CAUUCUGGCUGAAAUCCAAUCACGACCACAAUCUUUAGCUCAUAAUCAAACUUGAACUCUACACUUUUGCUACUCCUCCUCCAUCG .....(((((.((((((((((.....(((((....(((.(((((((.......))).).))).)))....)))))....(((....)))..............)))))))))).))))). (-25.74 = -27.80 + 2.06)

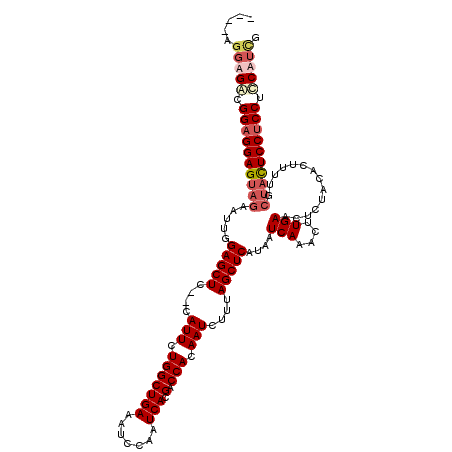

| Location | 16,258,147 – 16,258,265 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.52 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -34.51 |
| Energy contribution | -35.33 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16258147 118 - 22224390 CGAAGAAGGAGGAGUAGCAAAAGUGUAGAGUUCAAGUUUGAUUAUGAGCUAAAGAUUGUGGUCGUGAUUGGAUUUCAGCCAGAAUG--GAGCUCCAAUUCUACUCCUCCGUCUCCUCUUU .(.(((.((((((((((....(((...(((((((((((..(((((((.(..........).)))))))..)))))((.......))--))))))..))))))))))))).))).)..... ( -38.90) >DroSec_CAF1 6472 113 - 1 CGAAGGAGGAGGAGUAGCAAAAGUGUAGAGUUCAAGUUUGAUUAUGAGCUAAAGAUUGUGGUCGUGAUUGGAUUUCAGCCAGAAUGGGGAGCUCCAAUUCUACUCCUCCGUCU------- ...((..((((((((((....(((...(((((((((((..(((((((.(..........).)))))))..)))))...((.....)).))))))..)))))))))))))..))------- ( -40.60) >DroSim_CAF1 6372 117 - 1 CAAUGGUGGUGGAAU---UCAAGUGUAGAGUUCAAGUUUGAUUAUGAGCUAAAGAUUGUGGUCGUGAUUGGAUUUCAGCCAGAAUGGGGAGCUCCAAUUCUACUCCUCCGUCUCCUCUUU ..((((.((((((((---(........(((((((((((..(((((((.(..........).)))))))..)))))...((.....)).)))))).)))))))))...))))......... ( -30.10) >DroEre_CAF1 6561 114 - 1 GGAUGGAGGAGGAGUAGCAAAAGUGUAGAGUUCAAGUUUGAUUAUGAGCUAAAGAUUGUGGUCGUGAUUGGAUUUCAGCCAGAAUG--GAGCUCCACUUCUGCUCCUCCUCCUCCU---- (((.(((((((((((((...(((((..(((((((((((..(((((((.(..........).)))))))..)))))((.......))--))))))))))))))))))))))))))).---- ( -52.90) >consensus CGAAGGAGGAGGAGUAGCAAAAGUGUAGAGUUCAAGUUUGAUUAUGAGCUAAAGAUUGUGGUCGUGAUUGGAUUUCAGCCAGAAUG__GAGCUCCAAUUCUACUCCUCCGUCUCCU____ ....(((((((((((((......((..(((((((((((..(((((((.(..........).)))))))..)))))((.......))..))))))))...)))))))))))))........ (-34.51 = -35.33 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:50 2006