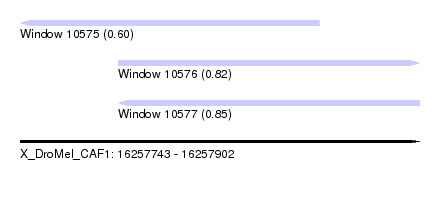
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,257,743 – 16,257,902 |
| Length | 159 |
| Max. P | 0.847888 |

| Location | 16,257,743 – 16,257,862 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.82 |
| Mean single sequence MFE | -36.66 |
| Consensus MFE | -33.50 |
| Energy contribution | -33.10 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16257743 119 - 22224390 CCUCACUUUUCCAUUCUGUCCAGCGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGACCAUUUGGGAUCUGUGUGUAUAACUGUUAAUGGUGUCCAACCACCAGAGCACCG-GAGUGC ...(((..(((((...((..(((.(((.(((((.(((....))).))))))))..)))..))..)))))((((.((((....(((....((((.....)))).))).))))))-))))). ( -35.80) >DroSec_CAF1 6068 119 - 1 CCUCACUUUUCCAUUCUGUCCAACGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGACCAUUUGGGAUCUGUGUGUAUAACUGUUAAUGGUGUCCAACCACCAGAGCACCG-GAGUGC ...(((..(((((....(((....(((.(((((.(((....))).))))))))....)))....)))))((((.((((....(((....((((.....)))).))).))))))-))))). ( -34.70) >DroSim_CAF1 5963 119 - 1 CCUCACUUUUCCAUUCUGUCCAGCGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGACCAUUUGGGAUCUGUGUGUAUAACUGUUAAUGGUGUCCAACCACCAGAGCACCG-GAGUGC ...(((..(((((...((..(((.(((.(((((.(((....))).))))))))..)))..))..)))))((((.((((....(((....((((.....)))).))).))))))-))))). ( -35.80) >DroEre_CAF1 6157 119 - 1 CCUCACUUUUCCGUUCUGUCCAGCGAACGGGUGUGGCCACUGCCUCAUCCUUCCACUGGCCAUUUGGGAUCUGUGUGUAUAACUGUUAAUGGUGUCCAACCACCAGAGCACCG-GAGUGC ...(((..(((((...((.((((.(((.(((((.(((....))).))))))))..)))).))..)))))((((.((((....(((....((((.....)))).))).))))))-))))). ( -37.90) >DroYak_CAF1 5981 120 - 1 CCUCACUUUUCCAUACUGUCCAGCGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGGCCAUUUGGGAUCUGUGUGUAUAACUGUUAAUGGUGUCCAACCACCAGAGCACCGGGAGUGC ...((((((((((...((.((((.(((.(((((.(((....))).))))))))..)))).))..))))).(((.((((....(((....((((.....)))).))).)))))))))))). ( -39.10) >consensus CCUCACUUUUCCAUUCUGUCCAGCGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGACCAUUUGGGAUCUGUGUGUAUAACUGUUAAUGGUGUCCAACCACCAGAGCACCG_GAGUGC .(.(((..(((((....(((....(((.(((((.(((....))).))))))))....)))....)))))...))).)............(((((......)))))..((((.....)))) (-33.50 = -33.10 + -0.40)

| Location | 16,257,782 – 16,257,902 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -39.39 |
| Consensus MFE | -32.78 |
| Energy contribution | -33.02 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16257782 120 + 22224390 AUACACACAGAUCCCAAAUGGUCAGUGGAAGGAUGAGGCGGUGGCCACACCCGUUCGCUGGACAGAAUGGAAAAGUGAGGUUACGGAUGAUGCCGAGUAACCAUUGGGCAGUGGGCAUUG .........(((((((..((.(((((((....((..((((((((((.(((((((((........))))))....))).))))))......))))..))..))))))).)).)))).))). ( -38.90) >DroSec_CAF1 6107 120 + 1 AUACACACAGAUCCCAAAUGGUCAGUGGAAGGAUGAGGCGGUGGCCACACCCGUUCGUUGGACAGAAUGGAAAAGUGAGGUUACGGAUGAUGUCGAUUAGCCAUUGGGCAGUGGGCAUUG .........(((((((..((.(((((((...(((..((((((((((.(((((((((........))))))....))).))))))......)))).)))..))))))).)).)))).))). ( -37.40) >DroSim_CAF1 6002 120 + 1 AUACACACAGAUCCCAAAUGGUCAGUGGAAGGAUGAGGCGGUGGCCACACCCGUUCGCUGGACAGAAUGGAAAAGUGAGGUUACGGAUGAUGCCGAGUAGCCAUUGGGCAGUGGGCAUUG .....(((...(((....((..(((((((.((.((.(((....))).)).)).)))))))..))....)))...)))...........((((((.(.(.(((....)))).).)))))). ( -40.80) >DroEre_CAF1 6196 120 + 1 AUACACACAGAUCCCAAAUGGCCAGUGGAAGGAUGAGGCAGUGGCCACACCCGUUCGCUGGACAGAACGGAAAAGUGAGGUUACGGAUGAUGUCGAGAAGCCAUGGGCCAUGGGGCAUUG .........(.(((((..(((((.((((........((((((((((.(((((((((........))))))....))).))))))......))))......)))).))))))))))).... ( -41.94) >DroYak_CAF1 6021 113 + 1 AUACACACAGAUCCCAAAUGGCCAGUGGAAGGAUGAGGCGGUGGCCACACCCGUUCGCUGGACAGUAUGGAAAAGUGAGGUUACGGAUGAUGCCGAGUAGCUAUUGG-------GCAUUG .....(((...(((....((.((((((((.((.((.(((....))).)).)).)))))))).))....)))...)))...........((((((.(((....))).)-------))))). ( -37.90) >consensus AUACACACAGAUCCCAAAUGGUCAGUGGAAGGAUGAGGCGGUGGCCACACCCGUUCGCUGGACAGAAUGGAAAAGUGAGGUUACGGAUGAUGCCGAGUAGCCAUUGGGCAGUGGGCAUUG .........((((((...((.(((((((........((((((((((.(((((((((........))))))....))).))))))......))))......))))))).))..))).))). (-32.78 = -33.02 + 0.24)

| Location | 16,257,782 – 16,257,902 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -33.19 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.70 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
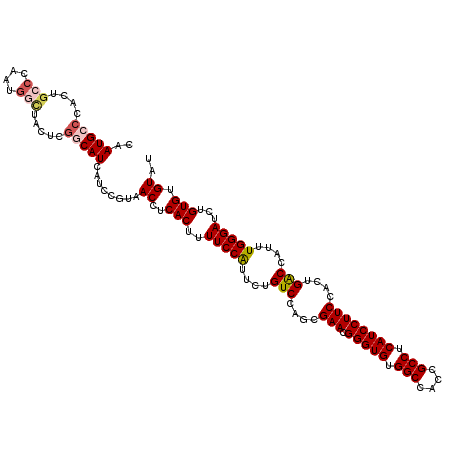
>X_DroMel_CAF1 16257782 120 - 22224390 CAAUGCCCACUGCCCAAUGGUUACUCGGCAUCAUCCGUAACCUCACUUUUCCAUUCUGUCCAGCGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGACCAUUUGGGAUCUGUGUGUAU ..((((.(((..(((((.((((((..((......))))))))..............((..(((.(((.(((((.(((....))).))))))))..)))..)).)))))....))).)))) ( -35.50) >DroSec_CAF1 6107 120 - 1 CAAUGCCCACUGCCCAAUGGCUAAUCGACAUCAUCCGUAACCUCACUUUUCCAUUCUGUCCAACGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGACCAUUUGGGAUCUGUGUGUAU ..((((.(((..((((((((......((((..........................))))....(((.(((((.(((....))).))))))))......))).)))))....))).)))) ( -29.47) >DroSim_CAF1 6002 120 - 1 CAAUGCCCACUGCCCAAUGGCUACUCGGCAUCAUCCGUAACCUCACUUUUCCAUUCUGUCCAGCGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGACCAUUUGGGAUCUGUGUGUAU ..(((((....(((....))).....)))))........((..(((..(((((...((..(((.(((.(((((.(((....))).))))))))..)))..))..)))))...))).)).. ( -35.20) >DroEre_CAF1 6196 120 - 1 CAAUGCCCCAUGGCCCAUGGCUUCUCGACAUCAUCCGUAACCUCACUUUUCCGUUCUGUCCAGCGAACGGGUGUGGCCACUGCCUCAUCCUUCCACUGGCCAUUUGGGAUCUGUGUGUAU ......(((((((((..(((................................((((........))))(((((.(((....))).)))))..)))..)))))..))))............ ( -33.20) >DroYak_CAF1 6021 113 - 1 CAAUGC-------CCAAUAGCUACUCGGCAUCAUCCGUAACCUCACUUUUCCAUACUGUCCAGCGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGGCCAUUUGGGAUCUGUGUGUAU ..((((-------(............)))))........((..(((..(((((...((.((((.(((.(((((.(((....))).))))))))..)))).))..)))))...))).)).. ( -32.60) >consensus CAAUGCCCACUGCCCAAUGGCUACUCGGCAUCAUCCGUAACCUCACUUUUCCAUUCUGUCCAGCGAACGGGUGUGGCCACCGCCUCAUCCUUCCACUGACCAUUUGGGAUCUGUGUGUAU ..(((((....(((....))).....)))))........((..(((..(((((....(((....(((.(((((.(((....))).))))))))....)))....)))))...))).)).. (-25.82 = -26.70 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:47 2006