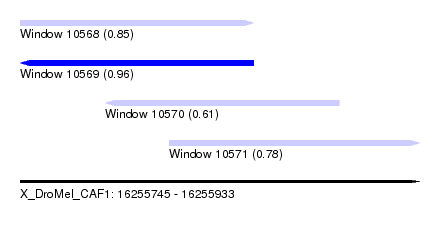
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,255,745 – 16,255,933 |
| Length | 188 |
| Max. P | 0.962603 |

| Location | 16,255,745 – 16,255,855 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -40.57 |
| Consensus MFE | -28.84 |
| Energy contribution | -30.65 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.850100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16255745 110 + 22224390 AAACGCACACAAGUGCAGCACAUCAACACGUGUCAGUGCCGGUGUGCGUGCGUGCGUGU----------GUGUGAGUAGUUGCAUUUCGAGAUACAAUUGUAUCUUUGUAUCUGAAGAUA ...((((((((.((((((((((((..(((......)))..))))))).)))))...)))----------)))))..........((((.((((((((........))))))))))))... ( -40.90) >DroSec_CAF1 4071 112 + 1 AAACGCAGACAAGUGCAGCACAUCAACACGUGUCAGUGCCGGUGUGCGUGCGUGCGUGUG--------UGUGUGAGUAGUUGCAUUUCGAGAUACAAUUGUAUCUUUGUAUCUUAAGAUA ..(((((.(((.((((((((((((..(((......)))..))))))).)))))...))).--------)))))..........((((..((((((((........))))))))..)))). ( -39.30) >DroSim_CAF1 3953 112 + 1 AAACGCAGACAAGUGCAGCACAUCAACACGUGUCAGUGCCGGUGUGCGUGCGUGCGUGUG--------UGUGUGAGCAGUUGCAUUUCGAGAUACAAUUGUAUCUUUGUAUCUUAAGAUA ..(((((.(((.((((((((((((..(((......)))..))))))).)))))...))).--------)))))..((....))((((..((((((((........))))))))..)))). ( -40.80) >DroYak_CAF1 3885 116 + 1 AAACGCACACAAGUGCAGCACAUCAACACGUGUCAGUGCCGGUUUGCG----UGUGUGUGCGUGUGCAUGUGUGGGUAGUUGCAUUUUGAGAUACAAUUGUAUCUUAGUAUCUGAAGAUA ..((.((((((.(..((((((((..(((((((............))))----))))))))).))..).)))))).)).........(((((((((....)))))))))((((....)))) ( -41.30) >consensus AAACGCACACAAGUGCAGCACAUCAACACGUGUCAGUGCCGGUGUGCGUGCGUGCGUGUG________UGUGUGAGUAGUUGCAUUUCGAGAUACAAUUGUAUCUUUGUAUCUGAAGAUA ...(((((.((.((((.(((((((..(((......)))..))))))))))).)).)))))...........(..(....)..)((((..((((((((........))))))))..)))). (-28.84 = -30.65 + 1.81)

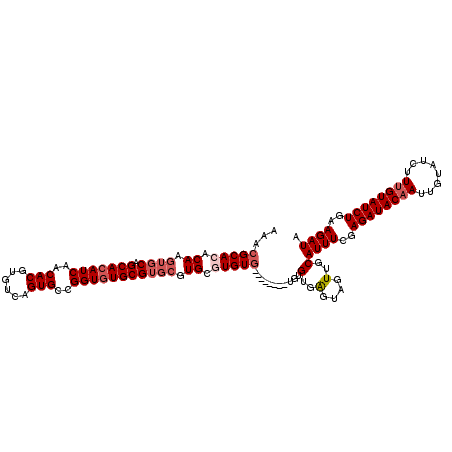
| Location | 16,255,745 – 16,255,855 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -25.06 |
| Energy contribution | -26.38 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16255745 110 - 22224390 UAUCUUCAGAUACAAAGAUACAAUUGUAUCUCGAAAUGCAACUACUCACAC----------ACACGCACGCACGCACACCGGCACUGACACGUGUUGAUGUGCUGCACUUGUGUGCGUUU ....(((((((((((........)))))))).)))............((.(----------(((((...(((.(((((.((((((......)))))).))))))))...)))))).)).. ( -35.80) >DroSec_CAF1 4071 112 - 1 UAUCUUAAGAUACAAAGAUACAAUUGUAUCUCGAAAUGCAACUACUCACACA--------CACACGCACGCACGCACACCGGCACUGACACGUGUUGAUGUGCUGCACUUGUCUGCGUUU ....(..((((((((........))))))))..).................(--------(.((.(((.(((.(((((.((((((......)))))).))))))))...))).)).)).. ( -29.00) >DroSim_CAF1 3953 112 - 1 UAUCUUAAGAUACAAAGAUACAAUUGUAUCUCGAAAUGCAACUGCUCACACA--------CACACGCACGCACGCACACCGGCACUGACACGUGUUGAUGUGCUGCACUUGUCUGCGUUU ....(..((((((((........))))))))..)(((((((((((.......--------.....))).(((.(((((.((((((......)))))).))))))))....)).)))))). ( -32.20) >DroYak_CAF1 3885 116 - 1 UAUCUUCAGAUACUAAGAUACAAUUGUAUCUCAAAAUGCAACUACCCACACAUGCACACGCACACACA----CGCAAACCGGCACUGACACGUGUUGAUGUGCUGCACUUGUGUGCGUUU ((((....))))...((((((....)))))).....((((............)))).(((((((((((----.(((...((((((......))))))...)))))....))))))))).. ( -31.10) >consensus UAUCUUAAGAUACAAAGAUACAAUUGUAUCUCGAAAUGCAACUACUCACACA________CACACGCACGCACGCACACCGGCACUGACACGUGUUGAUGUGCUGCACUUGUCUGCGUUU ((((....))))...((((((....))))))...............................((((((.(((.(((((.((((((......)))))).))))))))...))))))..... (-25.06 = -26.38 + 1.31)

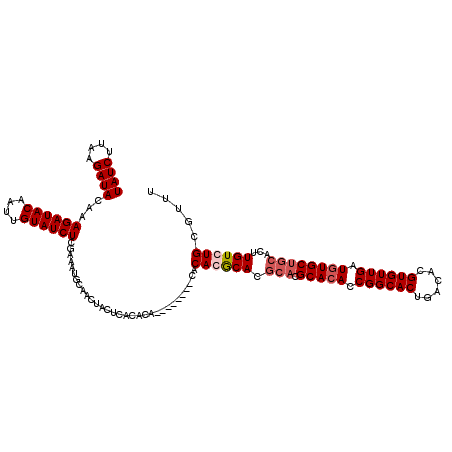

| Location | 16,255,785 – 16,255,895 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.22 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.03 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16255785 110 - 22224390 ACUGGUGGCGGGGUAACCAGCGAUAGAAACUUUCGUUUGAUAUCUUCAGAUACAAAGAUACAAUUGUAUCUCGAAAUGCAACUACUCACAC----------ACACGCACGCACGCACACC ...(((((((((....)).(((........(((((((((.((((....))))))))(((((....))))).)))))(((............----------....)))))).))).)))) ( -28.09) >DroSec_CAF1 4111 112 - 1 ACUGGCGGCGGGGUAACCAGCGAUAGAAACUUUCGUUUGGUAUCUUAAGAUACAAAGAUACAAUUGUAUCUCGAAAUGCAACUACUCACACA--------CACACGCACGCACGCACACC ....((((((((....)).(((....((((....)))).((((.(..((((((((........))))))))..).)))).............--------....))).))).)))..... ( -28.80) >DroSim_CAF1 3993 112 - 1 ACUGGCGGCGGGGUAACCAGCGAUAGAAACUUUCGUUUGGUAUCUUAAGAUACAAAGAUACAAUUGUAUCUCGAAAUGCAACUGCUCACACA--------CACACGCACGCACGCACACC ....((((((((....)).(((........(((((....(((((....)))))..((((((....))))))))))).((....)).......--------....))).))).)))..... ( -30.20) >DroYak_CAF1 3925 116 - 1 ACUGACAGCGGGGUAACCAGCAAUAGAAACAUUCGUUUGGUAUCUUCAGAUACUAAGAUACAAUUGUAUCUCAAAAUGCAACUACCCACACAUGCACACGCACACACA----CGCAAACC .......((((((((....(((..............((((((((....))))))))(((((....)))))......)))...))))).....(((....)))......----)))..... ( -28.70) >consensus ACUGGCGGCGGGGUAACCAGCGAUAGAAACUUUCGUUUGGUAUCUUAAGAUACAAAGAUACAAUUGUAUCUCGAAAUGCAACUACUCACACA________CACACGCACGCACGCACACC .......((((((((....(((...((.....)).(((((((((....)))))..((((((....)))))))))).)))...)))))..................(....).)))..... (-18.27 = -18.03 + -0.25)

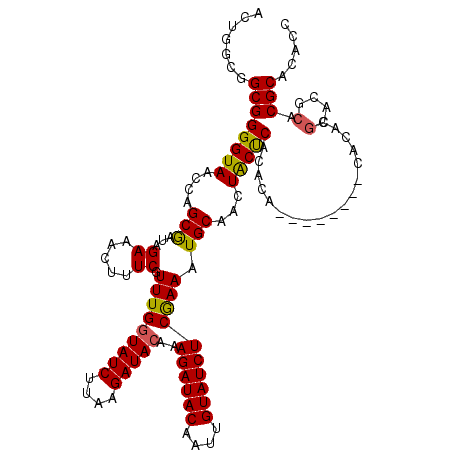
| Location | 16,255,815 – 16,255,933 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -26.94 |
| Consensus MFE | -21.34 |
| Energy contribution | -20.74 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16255815 118 + 22224390 UGCAUUUCGAGAUACAAUUGUAUCUUUGUAUCUGAAGAUAUCAAACGAAAGUUUCUAUCGCUGGUUACCCCGCCACCAGUCGUACGAUAUAUGUAUUUCUUCUCGGCAGAUACAC--ACA ........(((((((....)))))))((((((((..((((((((((....)))).(((.((((((.........)))))).))).)))).............))..)))))))).--... ( -29.41) >DroSec_CAF1 4143 118 + 1 UGCAUUUCGAGAUACAAUUGUAUCUUUGUAUCUUAAGAUACCAAACGAAAGUUUCUAUCGCUGGUUACCCCGCCGCCAGUCGUACGAUAUAUGUAUUUCUUCUCGGUAGAUACAC--ACA ....(((((((((((....))))))..(((((....)))))....)))))((.((((((((((((.........)))))).(((((.....)))))........)))))).))..--... ( -28.60) >DroSim_CAF1 4025 118 + 1 UGCAUUUCGAGAUACAAUUGUAUCUUUGUAUCUUAAGAUACCAAACGAAAGUUUCUAUCGCUGGUUACCCCGCCGCCAGUCGUACGAUAUAUGUAUUUCUUCUCGGUAGAUACAC--ACA ....(((((((((((....))))))..(((((....)))))....)))))((.((((((((((((.........)))))).(((((.....)))))........)))))).))..--... ( -28.60) >DroEre_CAF1 4049 118 + 1 UGCAUUUCAAGAUAUAAUUGUAUCGUUGUAUCUAAAGAUACCAAACGAAUGUUUCUAUUGCAGGUUACCCCGCCACCAGUCGUACGGUCUAUGUCUUUUUGCUCCGUAGAUACAC--ACA .(((....((((((((((((((((((((((((....)))))..)))))........((((..(((......)))..))))..)))))).))))))))..))).............--... ( -23.80) >DroYak_CAF1 3961 120 + 1 UGCAUUUUGAGAUACAAUUGUAUCUUAGUAUCUGAAGAUACCAAACGAAUGUUUCUAUUGCUGGUUACCCCGCUGUCAGUCGUACGAUAUAUAUAUUUUUGCUCGGUAGAUACACACACA .......((((((((....))))))))(((((((..((...((((.(.((((.(((((.(((((...........))))).))).))...)))).).)))).))..)))))))....... ( -24.30) >consensus UGCAUUUCGAGAUACAAUUGUAUCUUUGUAUCUGAAGAUACCAAACGAAAGUUUCUAUCGCUGGUUACCCCGCCGCCAGUCGUACGAUAUAUGUAUUUCUUCUCGGUAGAUACAC__ACA (((.....((((((((..((((((...(((((....))))).((((....)))).(((.((((((.........)))))).))).))))))))))))))......)))............ (-21.34 = -20.74 + -0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:41 2006