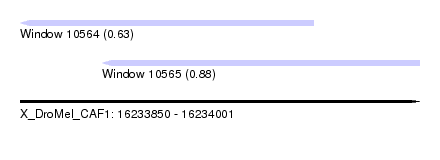
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,233,850 – 16,234,001 |
| Length | 151 |
| Max. P | 0.879810 |

| Location | 16,233,850 – 16,233,961 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -34.78 |
| Consensus MFE | -23.32 |
| Energy contribution | -24.52 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
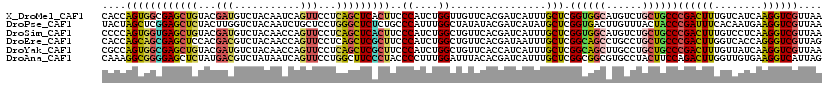
>X_DroMel_CAF1 16233850 111 - 22224390 AGCUCACUUCCCAUCUGGUUGUUCACGAUCAUUUGCUCGGUGGCAUGUCUGCUGCCCGACUUUGUCAUCAAGGUCGUUAAUGAUAUGUACAGGAAAG------UGCCAC---UCAAAAGC .(((.......((..((((((....))))))..))...((((((((.((((.(((.((((((((....))))))))..........)))))))...)------))))))---)....))) ( -31.60) >DroSec_CAF1 16 111 - 1 AGCUCACUUCCCAUCUGGUUGUUCACGAUCAUUUGCUCGGUGGCAUGUCUGCUGCCCGACUUUGUCAUCAAGGUCGUUAAUGAUAUGUACAGGAAAG------UGCCAC---CCAAGAGC .((((......((..((((((....))))))..))...((((((((.((((.(((.((((((((....))))))))..........)))))))...)------))))))---)...)))) ( -38.40) >DroSim_CAF1 52 111 - 1 AGCUCACUUCCCAUCUGGCUGUUCACGAUCAUUUGCUCGGUGGCAUGUCUGCUGCCCGACUUUGUCCUCAAGGUCGUUAAUGAUAUGUACAGGAAAG------UGCCAC---CCAAGAGC .((((......((..((..((....))..))..))...((((((((.((((.(((.((((((((....))))))))..........)))))))...)------))))))---)...)))) ( -35.00) >DroEre_CAF1 40 114 - 1 AGCUCGCUUCCCAUCUGGCUGUUCACGAUAAUUUGCUCGGCAGCCUGCCUGCUGCCCGACUUGGUCACCAGGGUCGUUAGUGACAUGUCCAGAAACG------UGCCACCAACCAAGAGC .((((........(((((((((.(((............((((((......))))))((((((((...))).)))))...)))))).).)))))...(------(.......))...)))) ( -34.40) >DroYak_CAF1 178 111 - 1 AGCUCGCUUCCCAUCUGGCUGUUCACCAUCAUUUGCUCGGCAGCUUGCCUGCUGCCCGACUUUGUUAUCAAGGUCGUUAAUGAUAUGUACAGGAAGG------UGCCAC---CCAAGAGU .((((......(((((..((((.((..((((((.((..((((((......)))))).(((((((....))))))))).)))))).)).))))..)))------))....---....)))) ( -40.14) >DroPer_CAF1 49 117 - 1 GGCUCUCUGCCCAUUUGGCUAUAUACGAUCAUAUGCUCGGUGACUUGUUUACUACCCGAUUUCACAAUGAAGGUCGUUAAGGAUAUGCUCAGGCAACCAAUGCCGCCAU---CCGACAUA (((.....)))....((((.......((.(((((.((..(((((((........................)))))))..)).))))).)).((((.....)))))))).---........ ( -29.16) >consensus AGCUCACUUCCCAUCUGGCUGUUCACGAUCAUUUGCUCGGUGGCAUGUCUGCUGCCCGACUUUGUCAUCAAGGUCGUUAAUGAUAUGUACAGGAAAG______UGCCAC___CCAAGAGC .((((.....((....))((((.((..((((((.((..((((((......)))))).(((((((....))))))))).)))))).)).))))........................)))) (-23.32 = -24.52 + 1.20)

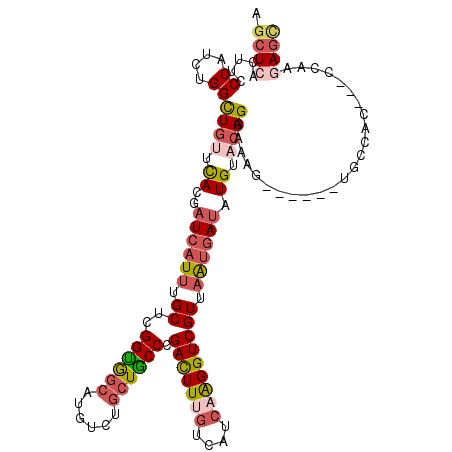
| Location | 16,233,881 – 16,234,001 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.56 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -24.62 |
| Energy contribution | -24.18 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16233881 120 - 22224390 CACCAGUGGCGAGCUGUACGAUGUCUACAAUCAGUUCCUCAGCUCACUUCCCAUCUGGUUGUUCACGAUCAUUUGCUCGGUGGCAUGUCUGCUGCCCGACUUUGUCAUCAAGGUCGUUAA .((((((((.((((((.(((((.......))).))....)))))).....))).)))))...................((..(((....)))..))((((((((....)))))))).... ( -37.20) >DroPse_CAF1 86 120 - 1 UACUAGCUCGGAGCUCUACUUGGUCUACAAUCUGCUCCUGGGCUCUCUGCCCAUUUGGCUAUAUACGAUCAUAUGCUCGGUGACUUGUUUACUACCCGAUUUCACAAUGAAGGUCGUUAA ...(((((.(((((.....(((.....)))...)))))(((((.....)))))...)))))...((((((......((((...............)))).(((.....)))))))))... ( -31.56) >DroSim_CAF1 83 120 - 1 CCCCAGUGGUGAGCUGUACGAUGUCUACAACCAGUUCCUCAGCUCACUUCCCAUCUGGCUGUUCACGAUCAUUUGCUCGGUGGCAUGUCUGCUGCCCGACUUUGUCCUCAAGGUCGUUAA ..((((.(((((((((...((...((......)).))..)))))))))......))))....................((..(((....)))..))((((((((....)))))))).... ( -37.00) >DroEre_CAF1 74 120 - 1 CACCAGCAGCGAGCUCCACGACGUCUACAACCAGUUCCUCAGCUCGCUUCCCAUCUGGCUGUUCACGAUAAUUUGCUCGGCAGCCUGCCUGCUGCCCGACUUGGUCACCAGGGUCGUUAG .....(((((((((.......(((..(((.((((......((....))......)))).)))..))).......))))(((.....))).))))).((((((((...))).))))).... ( -37.34) >DroYak_CAF1 209 120 - 1 CGCCAGUGGCGAGCUGUACGAUGUCUACAACCAGUUCCUCAGCUCGCUUCCCAUCUGGCUGUUCACCAUCAUUUGCUCGGCAGCUUGCCUGCUGCCCGACUUUGUUAUCAAGGUCGUUAA .(((((.(((((((((...((...((......)).))..)))))))))......)))))...................((((((......))))))((((((((....)))))))).... ( -45.70) >DroAna_CAF1 77 120 - 1 CAAAGGCGGGGAGCUCUAUGACGUCUAUAAUCAGUUCCUGGCUUCCCUACCCCUUUGGAUUUACACGAUCAUUUGCUCGGCGGCGUGCCUACUUCCAGACUUGGUUGUGAAGGUCAUUAG ((((((.((((((((....(((...........)))...))))))))....))))))(((((.((((((((....((.((.((........)).))))...)))))))).)))))..... ( -35.40) >consensus CACCAGCGGCGAGCUCUACGAUGUCUACAACCAGUUCCUCAGCUCACUUCCCAUCUGGCUGUUCACGAUCAUUUGCUCGGCGGCAUGCCUGCUGCCCGACUUUGUCAUCAAGGUCGUUAA ....((((((((((((...(((...........)))...)))))))))..((....))................))).((((((......))))))((((((........)))))).... (-24.62 = -24.18 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:36 2006