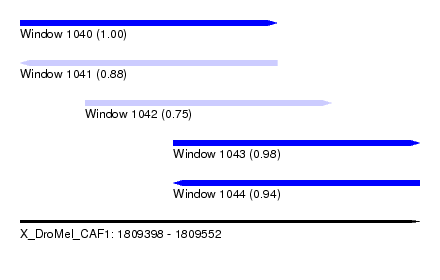
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,809,398 – 1,809,552 |
| Length | 154 |
| Max. P | 0.998542 |

| Location | 1,809,398 – 1,809,497 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.41 |
| Mean single sequence MFE | -16.59 |
| Consensus MFE | -14.63 |
| Energy contribution | -14.63 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1809398 99 + 22224390 ------UUGAAUUAACAAAACCCAUUUAAUUCCUUUCUUUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGC ------..(((((((..........)))))))....((((((((......((.....))..........)))))))).....((((((((......)))))))). ( -16.59) >DroSec_CAF1 17811 99 + 1 ------UUGAAUUAACAAAACCCAUUUGAUUCCUUUCUUUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGC ------..(((((((..........)))))))....((((((((......((.....))..........)))))))).....((((((((......)))))))). ( -16.69) >DroSim_CAF1 19761 99 + 1 ------UUGAAUUAACAAAACCCAUUUGAUUCCUUUCUUUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGC ------..(((((((..........)))))))....((((((((......((.....))..........)))))))).....((((((((......)))))))). ( -16.69) >DroEre_CAF1 17421 105 + 1 UUCGAAUUGAGCUAACCAACCCCACUUCAUUGCUUUCUUUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUACCAAAAUUGACAGC ..(((((((.(.....)............((((.......)))).......)))))))((((........))))........((((((((......)))))))). ( -16.30) >DroYak_CAF1 18539 105 + 1 UUCAAGUUGAGCUAACAAAAUUCACUUCAUUUCUUCCUUUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAAGAAAAUUGACAGC .....((((((....)....................((((((((......((.....))..........)))))))))))))((((((((......)))))))). ( -16.69) >consensus ______UUGAAUUAACAAAACCCAUUUCAUUCCUUUCUUUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGC ....................................((((((((......((.....))..........)))))))).....((((((((......)))))))). (-14.63 = -14.63 + -0.00)

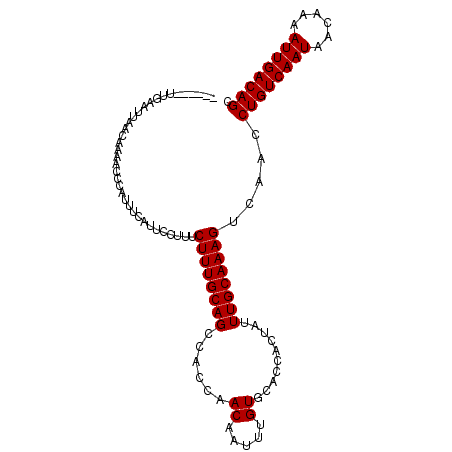
| Location | 1,809,398 – 1,809,497 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.41 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1809398 99 - 22224390 GCUGUCAAUUUUGUUAUUGACAGGUUGACUUUGCAAAUAGUGGUGCACAAAUUGUUGGUGGCUGCAAAGAAAGGAAUUAAAUGGGUUUUGUUAAUUCAA------ .((((((((......)))))))).....(((((((..(((..((......))..))).....)))))))....(((((((..........)))))))..------ ( -23.40) >DroSec_CAF1 17811 99 - 1 GCUGUCAAUUUUGUUAUUGACAGGUUGACUUUGCAAAUAGUGGUGCACAAAUUGUUGGUGGCUGCAAAGAAAGGAAUCAAAUGGGUUUUGUUAAUUCAA------ .((((((((......)))))))).(((((((((((..(((..((......))..))).....)))))))...((((((.....))))))......))))------ ( -23.00) >DroSim_CAF1 19761 99 - 1 GCUGUCAAUUUUGUUAUUGACAGGUUGACUUUGCAAAUAGUGGUGCACAAAUUGUUGGUGGCUGCAAAGAAAGGAAUCAAAUGGGUUUUGUUAAUUCAA------ .((((((((......)))))))).(((((((((((..(((..((......))..))).....)))))))...((((((.....))))))......))))------ ( -23.00) >DroEre_CAF1 17421 105 - 1 GCUGUCAAUUUUGGUAUUGACAGGUUGACUUUGCAAAUAGUGGUGCACAAAUUGUUGGUGGCUGCAAAGAAAGCAAUGAAGUGGGGUUGGUUAGCUCAAUUCGAA .((((((((......))))))))(((..(((((((..(((..((......))..))).....)))))))..)))..((((...((((......))))..)))).. ( -28.30) >DroYak_CAF1 18539 105 - 1 GCUGUCAAUUUUCUUAUUGACAGGUUGACUUUGCAAAUAGUGGUGCACAAAUUGUUGGUGGCUGCAAAGGAAGAAAUGAAGUGAAUUUUGUUAGCUCAACUUGAA .((((((((......))))))))((((((((((((..(((..((......))..))).....)))))))...(((((.......)))))......)))))..... ( -25.10) >consensus GCUGUCAAUUUUGUUAUUGACAGGUUGACUUUGCAAAUAGUGGUGCACAAAUUGUUGGUGGCUGCAAAGAAAGGAAUCAAAUGGGUUUUGUUAAUUCAA______ .((((((((......)))))))).....(((((((..(((..((......))..))).....)))))))............((((((.....))))))....... (-22.20 = -22.20 + -0.00)

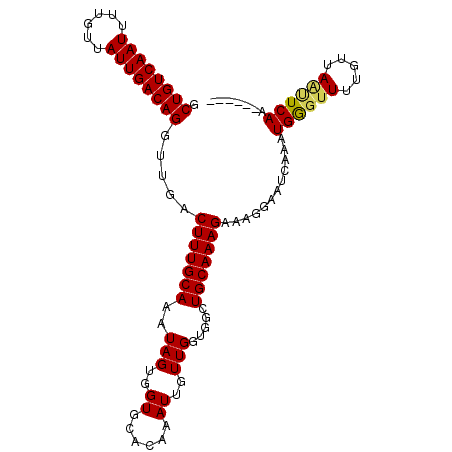
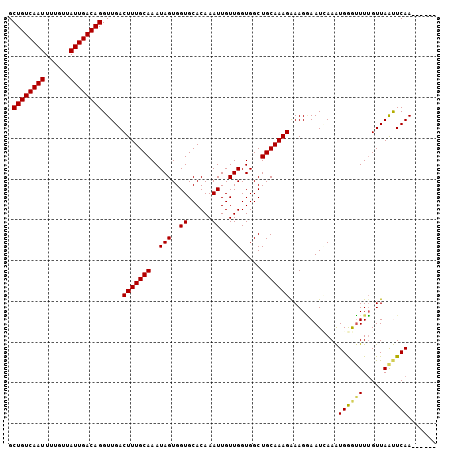
| Location | 1,809,423 – 1,809,518 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.36 |
| Mean single sequence MFE | -16.94 |
| Consensus MFE | -11.53 |
| Energy contribution | -12.20 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747428 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1809423 95 + 22224390 CCUUUCU-UUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGCUUUGAUAUUAAACAUAU----------------ACGU--- .....((-((((((......((.....))..........))))))))((((.((((((((......))))))))..))))............----------------....--- ( -16.59) >DroSec_CAF1 17836 95 + 1 CCUUUCU-UUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGCUUUGAUAUUAAACAUAU----------------ACGU--- .....((-((((((......((.....))..........))))))))((((.((((((((......))))))))..))))............----------------....--- ( -16.59) >DroSim_CAF1 19786 95 + 1 CCUUUCU-UUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGCUUUGAUAUUAAACAUAU----------------ACGU--- .....((-((((((......((.....))..........))))))))((((.((((((((......))))))))..))))............----------------....--- ( -16.59) >DroEre_CAF1 17452 90 + 1 GCUUUCU-UUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUACCAAAAUUGACAGCUUUGAUAUUAACCAUA------------------------ .....((-((((((......((.....))..........))))))))((((.((((((((......))))))))..))))...........------------------------ ( -16.59) >DroYak_CAF1 18570 95 + 1 UCUUCCU-UUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAAGAAAAUUGACAGCUUUGAUAUUAACCAUAU----------------ACGC--- .....((-((((((......((.....))..........))))))))((((.((((((((......))))))))..))))............----------------....--- ( -16.79) >DroAna_CAF1 21049 108 + 1 GUUUUCUAUUACAGCCACCAACAAUUUGUGCA------UUUAAAAGGUCAAAGUGUCAAUAAGAAUAUUGACAGCUUUGUU-UCAGACAUUUUCAAUCGAUCGGAUAUAGCUGAC ...........((((..((.....((((....------..)))).((((((((((((((((....)))))))).))))(((-...)))..........)))))).....)))).. ( -18.50) >consensus CCUUUCU_UUGCAGCCACCAACAAUUUGUGCACCACUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGCUUUGAUAUUAAACAUAU________________ACGU___ ............................((((........))))..(((((.((((((((......))))))))..))))).................................. (-11.53 = -12.20 + 0.67)

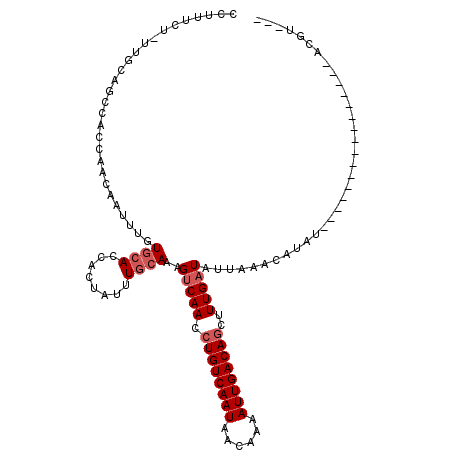
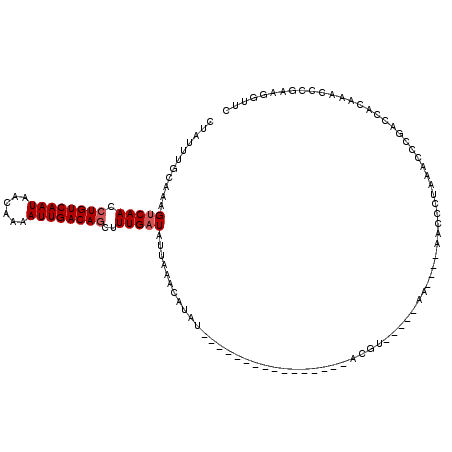
| Location | 1,809,457 – 1,809,552 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -15.28 |
| Consensus MFE | -10.47 |
| Energy contribution | -10.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1809457 95 + 22224390 CUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGCUUUGAUAUUAAACAUAU----------------ACGU-----AA----AACCCAAAACCCGACCACAAACCCAAAGUUUC ...........(((((.((((((((......))))))))..)))))...........----------------....-----..----................................ ( -12.30) >DroSec_CAF1 17870 95 + 1 CUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGCUUUGAUAUUAAACAUAU----------------ACGU-----AA----AACCCUAAAUCCGACCACAAACCCGAAGGUUC ...........(((((.((((((((......))))))))..)))))...........----------------....-----..----...................((((....)))). ( -14.00) >DroSim_CAF1 19820 95 + 1 CUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGCUUUGAUAUUAAACAUAU----------------ACGU-----AA----AACCCUAAACCCGACCACAAACCCGAAGGUUC ...........(((((.((((((((......))))))))..)))))...........----------------....-----..----...................((((....)))). ( -14.00) >DroEre_CAF1 17486 80 + 1 CUAUUUGCAAAGUCAACCUGUCAAUACCAAAAUUGACAGCUUUGAUAUUAACCAUA----------------------------------------ACCCGGCCACAAACCCGAAGGUUC ...........(((((.((((((((......))))))))..))))).........(----------------------------------------((((((........)))..)))). ( -16.50) >DroYak_CAF1 18604 95 + 1 CUAUUUGCAAAGUCAACCUGUCAAUAAGAAAAUUGACAGCUUUGAUAUUAACCAUAU----------------ACGC-----AA----CACCAUAAACCCGACCACAAGCCCGAAGGUUC ....((((...(((((.((((((((......))))))))..)))))((.......))----------------..))-----))----...................((((....)))). ( -14.40) >DroAna_CAF1 21081 116 + 1 ---UUUAAAAGGUCAAAGUGUCAAUAAGAAUAUUGACAGCUUUGUU-UCAGACAUUUUCAAUCGAUCGGAUAUAGCUGACAGAAGAUCGACUAUAAACCCGACAGCAAAUAAAAAAGUUC ---.......((.(((((((((((((....)))))))).)))))..-))..............(.((((.(((((.((((....).))).)))))...)))))................. ( -20.50) >consensus CUAUUUGCAAAGUCAACCUGUCAAUAACAAAAUUGACAGCUUUGAUAUUAAACAUAU________________ACGU_____AA____AACCCUAAACCCGACCACAAACCCGAAGGUUC ...........(((((.((((((((......))))))))..))))).......................................................................... (-10.47 = -10.80 + 0.33)

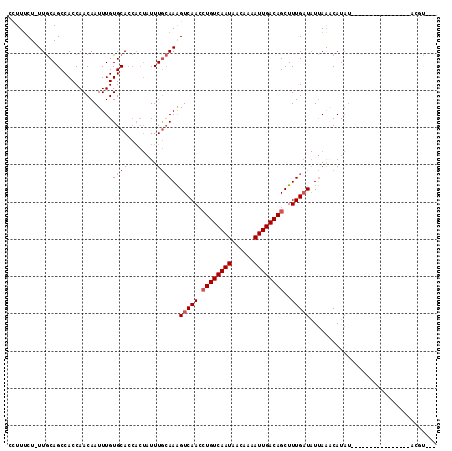
| Location | 1,809,457 – 1,809,552 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -24.65 |
| Consensus MFE | -13.82 |
| Energy contribution | -14.77 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
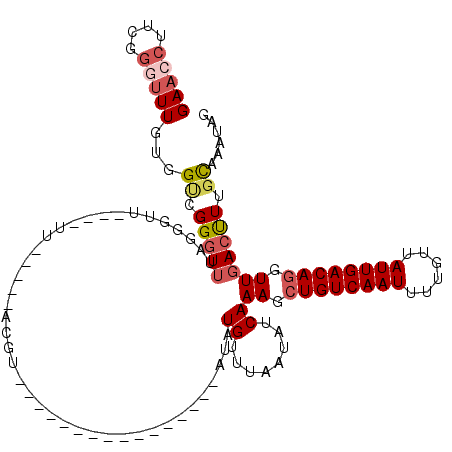
>X_DroMel_CAF1 1809457 95 - 22224390 GAAACUUUGGGUUUGUGGUCGGGUUUUGGGUU----UU-----ACGU----------------AUAUGUUUAAUAUCAAAGCUGUCAAUUUUGUUAUUGACAGGUUGACUUUGCAAAUAG ..........((((((((((((((((((((((----..-----(((.----------------...)))..))).))))))))((((((......))))))....)))))..)))))).. ( -21.80) >DroSec_CAF1 17870 95 - 1 GAACCUUCGGGUUUGUGGUCGGAUUUAGGGUU----UU-----ACGU----------------AUAUGUUUAAUAUCAAAGCUGUCAAUUUUGUUAUUGACAGGUUGACUUUGCAAAUAG (((((((.(((((((....)))))))))))))----).-----...(----------------((.(((......((((..((((((((......)))))))).))))....))).))). ( -22.40) >DroSim_CAF1 19820 95 - 1 GAACCUUCGGGUUUGUGGUCGGGUUUAGGGUU----UU-----ACGU----------------AUAUGUUUAAUAUCAAAGCUGUCAAUUUUGUUAUUGACAGGUUGACUUUGCAAAUAG (((((....)))))...((..((((....(((----..-----(((.----------------...)))..)))....((.((((((((......)))))))).))))))..))...... ( -22.20) >DroEre_CAF1 17486 80 - 1 GAACCUUCGGGUUUGUGGCCGGGU----------------------------------------UAUGGUUAAUAUCAAAGCUGUCAAUUUUGGUAUUGACAGGUUGACUUUGCAAAUAG (((((....)))))...((..(((----------------------------------------((..(((((((((((((.......)))))))))))))....)))))..))...... ( -26.00) >DroYak_CAF1 18604 95 - 1 GAACCUUCGGGCUUGUGGUCGGGUUUAUGGUG----UU-----GCGU----------------AUAUGGUUAAUAUCAAAGCUGUCAAUUUUCUUAUUGACAGGUUGACUUUGCAAAUAG ...((....)).((((((((.......(((((----((-----((..----------------.....).))))))))((.((((((((......)))))))).))))))..)))).... ( -24.30) >DroAna_CAF1 21081 116 - 1 GAACUUUUUUAUUUGCUGUCGGGUUUAUAGUCGAUCUUCUGUCAGCUAUAUCCGAUCGAUUGAAAAUGUCUGA-AACAAAGCUGUCAAUAUUCUUAUUGACACUUUGACCUUUUAAA--- ((...((((((.(((..(((((((..(((((.(((.....))).))))))))))))))).))))))..))...-..(((((.((((((((....)))))))))))))..........--- ( -31.20) >consensus GAACCUUCGGGUUUGUGGUCGGGUUUAGGGUU____UU_____ACGU________________AUAUGUUUAAUAUCAAAGCUGUCAAUUUUGUUAUUGACAGGUUGACUUUGCAAAUAG (((((....)))))...((.(((((.........................................((........))((.((((((((......)))))))).))))))).))...... (-13.82 = -14.77 + 0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:13 2006