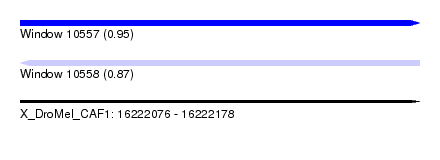
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,222,076 – 16,222,178 |
| Length | 102 |
| Max. P | 0.949957 |

| Location | 16,222,076 – 16,222,178 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -23.37 |
| Energy contribution | -23.99 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
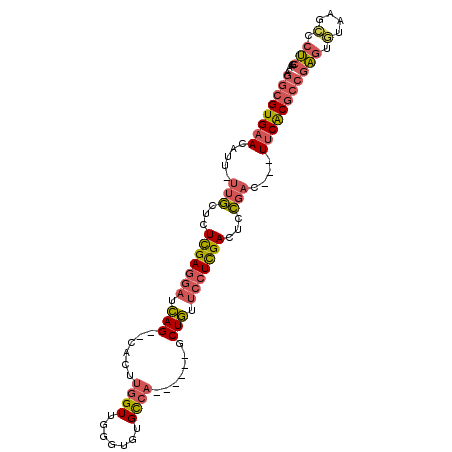
>X_DroMel_CAF1 16222076 102 + 22224390 AGGCGUGAACAUUUUUGCUCUCGAGGAUCAGCGCACUUGGUUGGGUGUGCCAGCA---GCUGUUCCUCGACUCCGACUCCUUCACGCCGAGUGUAAGCCCUCGAA .((((((((.....(((...(((((((.((((.......(((((.....))))).---)))).)))))))...)))....))))))))(((.(....).)))... ( -39.70) >DroSec_CAF1 1164 101 + 1 AGGCGUGAACAUUUUUGCUCUCGAGGAUCAG--CACUUGGUUGGGUGUGCCAGCAGCAGCUGUA-CUCGACU-CGACUCCUUCACGACGAGUGUAAGCCAUCGAA .((((....)....((((...((((...(((--(.....(((((.....)))))....))))..-))))(((-((.(........).))))))))))))...... ( -29.60) >DroSim_CAF1 1161 103 + 1 AGGCGUGAACAUUUUUGCUCUCGAGGAUCAG--CACUUGGUUGGGUGUGCCAGCAGCAGCUGUUCCUCGACUCCGACUCCUUCACGCCGAGUGUAAGCCCUCGAA .((((((((.....(((...(((((((.(((--(.....(((((.....)))))....)))).)))))))...)))....))))))))(((.(....).)))... ( -41.00) >DroEre_CAF1 1157 92 + 1 AGGCGUGAAAGU--UUGCUCUUGAGGAUCAG--CACUUGGUUGGGUGUGCCA------GCUGUUCCUCGACUCCGAC---UUCACGCCGAGUAUAAGCCCUCGAA .((((((((.((--(.(...(((((((.(((--(...((((.......))))------)))).)))))))..).)))---))))))))(((........)))... ( -36.80) >DroYak_CAF1 1146 92 + 1 AGGCGUGAAAGU--UUGCUCUUGAGGAUCAG--CACUUGGUUGGGUGUGCCA------GCUGUUCCUCGACUCCGAU---UUCACGCCGAGUGUAAGCCCUCGAA .(((((((((..--(((...(((((((.(((--(...((((.......))))------)))).)))))))...))))---))))))))(((.(....).)))... ( -36.90) >DroAna_CAF1 1104 86 + 1 GAGCGUGCACAUC---AUUCUUGAGGACUAG--CA----GUGGGGUGUGUCU------GCUAGUCAUUGAUUUUGAU---UUCGCACG-GGACCAUGGCCCCUAU ....((((..(((---(...(..(.((((((--((----(..........))------))))))).)..)...))))---...))))(-((.(....).)))... ( -29.10) >consensus AGGCGUGAACAUU_UUGCUCUCGAGGAUCAG__CACUUGGUUGGGUGUGCCA______GCUGUUCCUCGACUCCGAC___UUCACGCCGAGUGUAAGCCCUCGAA .((((((((.....(((...(((((((.(((......((((.......)))).......))).)))))))...)))....))))))))(((.(....).)))... (-23.37 = -23.99 + 0.62)


| Location | 16,222,076 – 16,222,178 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.96 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -16.44 |
| Energy contribution | -17.91 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
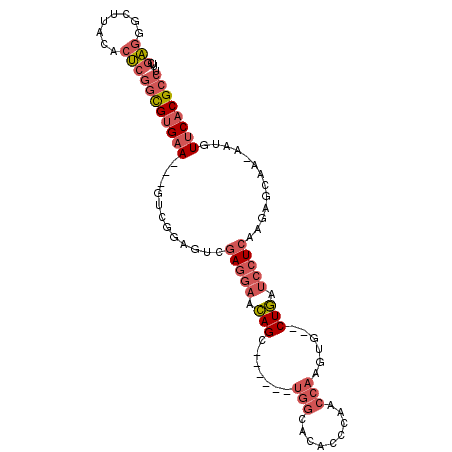
>X_DroMel_CAF1 16222076 102 - 22224390 UUCGAGGGCUUACACUCGGCGUGAAGGAGUCGGAGUCGAGGAACAGC---UGCUGGCACACCCAACCAAGUGCGCUGAUCCUCGAGAGCAAAAAUGUUCACGCCU ...(((........)))((((((((.(..(..(..(((((((.((((---((((((.........)).)))).)))).)))))))...)..)..).)))))))). ( -38.30) >DroSec_CAF1 1164 101 - 1 UUCGAUGGCUUACACUCGUCGUGAAGGAGUCG-AGUCGAG-UACAGCUGCUGCUGGCACACCCAACCAAGUG--CUGAUCCUCGAGAGCAAAAAUGUUCACGCCU ((((((..(((.(((.....))))))..))))-))(((((-..((((....))))((((..........)))--).....)))))(((((....)))))...... ( -30.30) >DroSim_CAF1 1161 103 - 1 UUCGAGGGCUUACACUCGGCGUGAAGGAGUCGGAGUCGAGGAACAGCUGCUGCUGGCACACCCAACCAAGUG--CUGAUCCUCGAGAGCAAAAAUGUUCACGCCU ...(((........)))((((((((.(..(..(..(((((((.((((.(((..(((.....)))....))))--))).)))))))...)..)..).)))))))). ( -38.80) >DroEre_CAF1 1157 92 - 1 UUCGAGGGCUUAUACUCGGCGUGAA---GUCGGAGUCGAGGAACAGC------UGGCACACCCAACCAAGUG--CUGAUCCUCAAGAGCAA--ACUUUCACGCCU ...(((........)))((((((((---((..(....(((((.((((------(((.........)))...)--))).))))).....)..--)).)))))))). ( -32.70) >DroYak_CAF1 1146 92 - 1 UUCGAGGGCUUACACUCGGCGUGAA---AUCGGAGUCGAGGAACAGC------UGGCACACCCAACCAAGUG--CUGAUCCUCAAGAGCAA--ACUUUCACGCCU ...(((........)))((((((((---(...(....(((((.((((------(((.........)))...)--))).))))).....)..--..))))))))). ( -31.40) >DroAna_CAF1 1104 86 - 1 AUAGGGGCCAUGGUCC-CGUGCGAA---AUCAAAAUCAAUGACUAGC------AGACACACCCCAC----UG--CUAGUCCUCAAGAAU---GAUGUGCACGCUC ...(((((....))))-)(((((..---((((........(((((((------((..........)----))--))))))........)---))).))))).... ( -31.89) >consensus UUCGAGGGCUUACACUCGGCGUGAA___GUCGGAGUCGAGGAACAGC______UGGCACACCCAACCAAGUG__CUGAUCCUCAAGAGCAA_AAUGUUCACGCCU ...(((........)))((((((((............(((((.(((.......(((.........)))......))).))))).............)))))))). (-16.44 = -17.91 + 1.47)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:28 2006