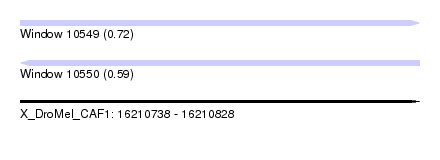
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,210,738 – 16,210,828 |
| Length | 90 |
| Max. P | 0.720644 |

| Location | 16,210,738 – 16,210,828 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.00 |
| Mean single sequence MFE | -14.88 |
| Consensus MFE | -12.01 |
| Energy contribution | -12.18 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16210738 90 + 22224390 AAAAAAAAAUCAAUGA----------UCAAUUGUUAAAAAUACUAUUUAAAC----GCAGGAUCCCGAGGCCUCUUUCGAUAUAAAUGGCAACGAUUCGGAUCC ..........((((..----------...))))...................----...((((((((..(((...............)))..))....)))))) ( -13.86) >DroSec_CAF1 865 89 + 1 AAAAA-AAUCCAAUGA----------UCAAUUGUUCAAAAUACUAUUUAAAC----GCAGGAUCCCGAGGCCUCUUUUGAUAUUAAUGGCAACGAUUCGGAUCC .....-.((((...((----------((...((((.(((......))).)))----)...)))).((..(((...............)))..))....)))).. ( -14.26) >DroSim_CAF1 852 89 + 1 AAAAA-AAUCCUAUGA----------UCAAUUGUUAAAAAUACGAUUUAAAC----GCAGGAUCCCGAGGCCUCUUUCGAUAUUAAUGGCAACGAUUCGGAUCC .....-..........----------..((((((.......)))))).....----...((((((((..(((...............)))..))....)))))) ( -14.96) >DroEre_CAF1 837 88 + 1 AAAAA-UAUCAAAACA----------CCAAAUGUA-AAAACACUAUUUAAAC----ACAGGAUCCCGAAGCCUCUUUCGACAUAAAUGGCAACGAUUCGGAUCC .....-..........----------.........-................----...(((((((((((....))))).......((....))....)))))) ( -12.80) >DroYak_CAF1 877 84 + 1 AAAAA-AGUCAA--------------UUAAUUGUU-AAAAAACUAUUUAAAC----ACAGGACCCCGAAGCCUCUUUCGACAUAAAUGGCAACGAUUCGGAUCC .....-......--------------.....((((-.(((.....))).)))----)..(((..((((((((...............))).....))))).))) ( -9.86) >DroPer_CAF1 1772 99 + 1 --AAA-AGUGCUAUGUAAAAUAUGCUCUAA-CGG-AAACAUACUCUUUCCGCAUUUUCAGGAUCCCGAUGCCUCAUUCGAUAUCAAUGGCAACGAUUCGGAUCC --.((-(((((...(((.....))).....-.((-(((.......))))))))))))..((((((((.((((...............)))).))....)))))) ( -23.56) >consensus AAAAA_AAUCCAAUGA__________UCAAUUGUUAAAAAUACUAUUUAAAC____GCAGGAUCCCGAGGCCUCUUUCGAUAUAAAUGGCAACGAUUCGGAUCC ...........................................................((((((((..(((...............)))..))....)))))) (-12.01 = -12.18 + 0.17)


| Location | 16,210,738 – 16,210,828 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.00 |
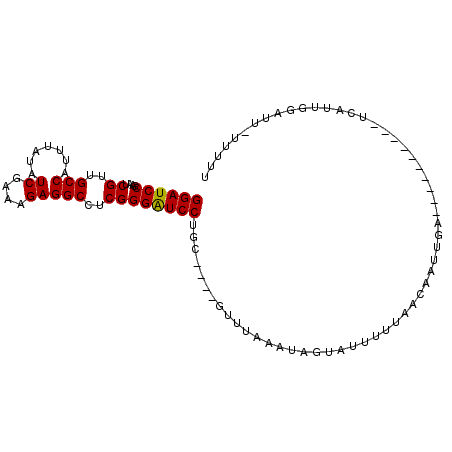
| Mean single sequence MFE | -20.03 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.08 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16210738 90 - 22224390 GGAUCCGAAUCGUUGCCAUUUAUAUCGAAAGAGGCCUCGGGAUCCUGC----GUUUAAAUAGUAUUUUUAACAAUUGA----------UCAUUGAUUUUUUUUU ((((((....((..(((.......((....)))))..))))))))...----(((.(((......))).)))......----------................ ( -16.71) >DroSec_CAF1 865 89 - 1 GGAUCCGAAUCGUUGCCAUUAAUAUCAAAAGAGGCCUCGGGAUCCUGC----GUUUAAAUAGUAUUUUGAACAAUUGA----------UCAUUGGAUU-UUUUU ((((((((.(((..(((...............)))..)))((((....----(((((((......)))))))....))----------)).)))))))-).... ( -20.66) >DroSim_CAF1 852 89 - 1 GGAUCCGAAUCGUUGCCAUUAAUAUCGAAAGAGGCCUCGGGAUCCUGC----GUUUAAAUCGUAUUUUUAACAAUUGA----------UCAUAGGAUU-UUUUU ((((((....((..(((.......((....)))))..))))))))...----....(((((.(((..(((.....)))----------..))).))))-).... ( -19.71) >DroEre_CAF1 837 88 - 1 GGAUCCGAAUCGUUGCCAUUUAUGUCGAAAGAGGCUUCGGGAUCCUGU----GUUUAAAUAGUGUUUU-UACAUUUGG----------UGUUUUGAUA-UUUUU ((((((....((..(((.......((....)))))..)))))))).((----(((.(((((((((...-.))))....----------))))).))))-).... ( -19.21) >DroYak_CAF1 877 84 - 1 GGAUCCGAAUCGUUGCCAUUUAUGUCGAAAGAGGCUUCGGGGUCCUGU----GUUUAAAUAGUUUUUU-AACAAUUAA--------------UUGACU-UUUUU ((((((....((..(((.......((....)))))..))))))))...----....(((.((((..((-(.....)))--------------..))))-.))). ( -16.71) >DroPer_CAF1 1772 99 - 1 GGAUCCGAAUCGUUGCCAUUGAUAUCGAAUGAGGCAUCGGGAUCCUGAAAAUGCGGAAAGAGUAUGUUU-CCG-UUAGAGCAUAUUUUACAUAGCACU-UUU-- ((((((....((.((((.(((....)))....)))).)))))))).((((.(((....(((((((((((-...-...))))))))))).....))).)-)))-- ( -27.20) >consensus GGAUCCGAAUCGUUGCCAUUUAUAUCGAAAGAGGCCUCGGGAUCCUGC____GUUUAAAUAGUAUUUUUAACAAUUGA__________UCAUUGGAUU_UUUUU ((((((....((..(((.......((....)))))..))))))))........................................................... (-14.22 = -14.08 + -0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:21 2006