| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,210,232 – 16,210,328 |
| Length | 96 |
| Max. P | 0.757717 |

| Location | 16,210,232 – 16,210,328 |
|---|---|
| Length | 96 |
| Sequences | 6 |
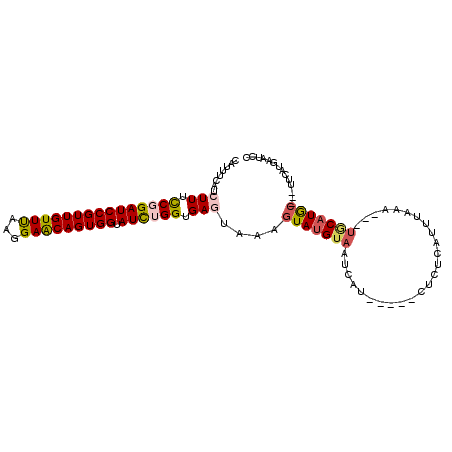
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -14.41 |
| Energy contribution | -14.69 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16210232 96 + 22224390 CAUUUCAUCUUUCCGGAUCCGUUGUUUAAGGAACAGUGGUAUCUGGUGAGUAAAGUAUGUAAUCAU-----CUCUCAUUUAAA---UGCAUGC---UUCAUGAAUCC ...((((((((.(((((((((((((((...))))))))).)))))).)))..(((((((((.....-----............---)))))))---)).)))))... ( -24.63) >DroSec_CAF1 372 96 + 1 CAUUUCAUCUUUCCGGAUCCGUUGUUUAAGGAACAGUGGUAUCUGGUGAGUAAAGUAUGUAAUCAU-----CACUCAUUUAAA---UGCAUGC---UUCAUGAAUCC ...((((((((.(((((((((((((((...))))))))).)))))).)))..(((((((((.....-----............---)))))))---)).)))))... ( -24.63) >DroSim_CAF1 374 96 + 1 CAUUUCAUCUUUCCGGAUCCGUUGUUUAAGGAACAGUGGUAUCUGGUGAGUAAAGUAUGUAAUCAU-----CUCUCAUUUAAA---UGCAUGC---UUCAUGAAUCC ...((((((((.(((((((((((((((...))))))))).)))))).)))..(((((((((.....-----............---)))))))---)).)))))... ( -24.63) >DroWil_CAF1 430 96 + 1 CAUUUAAUAUUCUCCGAUCCGUUGUUUAAGGAACAGUGGUAUUUGGUGAGUAAAGUAUGUAAUCGA-----UGUCGAUUUCAA---UGCAUGCAA-UUCA--AUUCA .......(((((.((((.(((((((((...)))))))))...)))).)))))..(((((((((((.-----...)))).....---)))))))..-....--..... ( -24.40) >DroMoj_CAF1 481 107 + 1 CAUCUCAUAUUCUCCGAUCCGUUGUUUAAGGAACAGUGGUAUUUGGUGAGUAAAGUAUGUCAUCAGGCCAUAUUUCAUCUCAACUAUACAUACACAUACAUAUAUCU .......(((((.((((.(((((((((...)))))))))...)))).)))))..((((((.((.((.................)))))))))).............. ( -19.23) >DroAna_CAF1 364 96 + 1 CACUCCAUCUUUCCGGAUCCGUUGUUCAAGGAGCAGUGGUAUCUGGUGAGUAAAGUAUGUAAUCAU-----AUCUCACUUAAA---UUCUUAA---UUUUUAAAACC ............(((((((((((((((...))))))))).))))))(((((.(..((((....)))-----)..).)))))..---.......---........... ( -21.10) >consensus CAUUUCAUCUUUCCGGAUCCGUUGUUUAAGGAACAGUGGUAUCUGGUGAGUAAAGUAUGUAAUCAU_____CUCUCAUUUAAA___UGCAUGC___UUCAUGAAUCC ........(((.(((((((((((((((...))))))))).)))))).)))....(((((((.........................))))))).............. (-14.41 = -14.69 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:18 2006