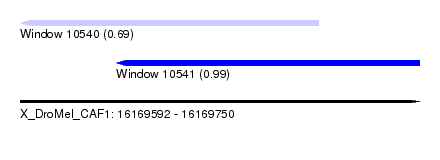
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,169,592 – 16,169,750 |
| Length | 158 |
| Max. P | 0.994337 |

| Location | 16,169,592 – 16,169,710 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -16.28 |
| Consensus MFE | -14.20 |
| Energy contribution | -14.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16169592 118 - 22224390 AUUAUUUACUUUCCAUGUAAAUAAAUAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUACCACACAAUAUAAUACUAAAUCCUGUCCAAAGUGAAUUCUCUUGUU .((((((((.......))))))))...((((((((.....))))))))((((.(((....))).))))........................((((......)).))........... ( -16.60) >DroSec_CAF1 9571 116 - 1 UUUAUUUACUUUCCAUGUAAAUAAAUAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUAACAC--ACUAUAAUACUAAAUCCUAUCCAAAGUGAAUUCUCUUGUU (((((((((.......)))))))))..((((((((.....))))))))((((.(((....))).)))).......(--(((.....................))))............ ( -17.60) >DroSim_CAF1 9948 116 - 1 UUUAUUUACUUUCCAUGUAAAUAAAUAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUAACAC--ACUAUAAUACUAAAUCCUAUCCACAGUGAAUUCUCUUGUU (((((((((.......)))))))))..((((((((.....))))))))((((.(((....))).)))).......(--(((.....................))))............ ( -17.40) >DroEre_CAF1 9992 116 - 1 UUCAUUUACUUUCCAUGUAAACAAACAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUACCAG--AAUAUAAUACUAAAUCCUGGCCAAAAUAAAGUCUCUUAUU ....(((((.......)))))......((((((((.....))))))))((((.(((....))).))))....((((--.((.((....)).)).)))).................... ( -15.30) >DroYak_CAF1 10573 116 - 1 UUCAUUUACUUUCCAUGUAAAUAAACGUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUACCAG--AAUAUAAUACCAAAUCCUGACCAAAAGAUAGUCUCUUAUU (((((((((.......)))))).....((((((((.....))))))))((((.(((....))).)))).......)--))......................((((.....))))... ( -14.50) >consensus UUUAUUUACUUUCCAUGUAAAUAAAUAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUACCAC__AAUAUAAUACUAAAUCCUGUCCAAAGUGAAUUCUCUUGUU .((((((((.......))))))))...((((((((.....))))))))((((.(((....))).)))).................................................. (-14.20 = -14.80 + 0.60)


| Location | 16,169,630 – 16,169,750 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -23.44 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.78 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16169630 120 - 22224390 AAAAAAAAAAAGUAUCUGUCGGAACUGGGGCGUUAUCCUUAUUAUUUACUUUCCAUGUAAAUAAAUAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUACCACACAA ................((((((...(((((......)))))((((((((.......))))))))...((((((((.....))))))))....)))))).((((....))))......... ( -23.80) >DroSec_CAF1 9609 110 - 1 A--------AAGUAUCUGUCGGAACUGGGGCGUUAUCCUUUUUAUUUACUUUCCAUGUAAAUAAAUAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUAACAC--AC .--------..((((((((.(((((.(((...((((....(((((((((.......)))))))))...))))...)))))).)).)))))))).((((......))))........--.. ( -24.30) >DroSim_CAF1 9986 110 - 1 A--------AAGUAUCUGUCGGAACUGGGGCGUUAUCCUUUUUAUUUACUUUCCAUGUAAAUAAAUAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUAACAC--AC .--------..((((((((.(((((.(((...((((....(((((((((.......)))))))))...))))...)))))).)).)))))))).((((......))))........--.. ( -24.30) >DroEre_CAF1 10030 110 - 1 A--------AAGUAUCUGUCGGAAUUGGGGCGUUAUCCUUUUCAUUUACUUUCCAUGUAAACAAACAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUACCAG--AA .--------..(((((((...(((..(((......)))..)))..........(((((......))))))))))))(((.((((....)))).)))...((((....)))).....--.. ( -21.80) >DroYak_CAF1 10611 110 - 1 A--------AAGUAUCUGUCGGAAUUGGGGCGUUAUCCUUUUCAUUUACUUUCCAUGUAAAUAAACGUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUACCAG--AA .--------..((((((((.((((..(((......)))..)))((((((.......)))))).((((.((....)).))))..).)))))))).((((......))))........--.. ( -23.00) >consensus A________AAGUAUCUGUCGGAACUGGGGCGUUAUCCUUUUUAUUUACUUUCCAUGUAAAUAAAUAUGUAGAUACUCGUUAUCUACAGAUACCGAUAAAUUGAUGUCAAUACCAC__AA ...........((((((((.(((..((((...((((....(((((((((.......)))))))))...))))...))))...))))))))))).((((......))))............ (-21.98 = -22.78 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:12 2006