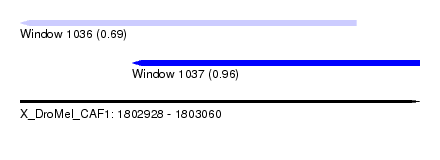
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 1,802,928 – 1,803,060 |
| Length | 132 |
| Max. P | 0.961017 |

| Location | 1,802,928 – 1,803,039 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.02 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -12.80 |
| Energy contribution | -13.44 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
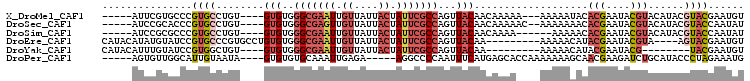
>X_DroMel_CAF1 1802928 111 - 22224390 UGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAACAAAAA-AAAAAUACACGAAUACGUACAUACGUACGAAUGUGUACAAAGCCAACAAAUUAAAAGAGAAACCCGAACUU ..(((((((.((....)).)))))))..............-.....((((((....(((((....)))))..)))))).................................. ( -21.30) >DroSec_CAF1 11438 112 - 1 UGGGCGAGUUGUUAUUACUAUUCGCCAGUUACAACAAAAACAAAAAAACACGAAUACGUACAUACGUACCAAUAUGUACAAAGCCAACAAAUUCAAAGAGAAACCCGAACUU ..(((((((.((....)).)))))))((((.(.........................(((((((........)))))))............(((.....)))....))))). ( -19.10) >DroSim_CAF1 11518 108 - 1 UGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAACAAAA----AAAAACACGAAUACGUACAUACGUACCAAUAUGUACAAAGCCAACAAAUUCAAAGAGAAACCCGAACUU ..(((((((.((....)).)))))))((((.(.......----..............(((((((........)))))))............(((.....)))....))))). ( -19.10) >DroEre_CAF1 11270 101 - 1 UGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAA-------AAAAACAUACGAAUACGUA----AGUACGAAUGUGCACAAAGUCAACAAAUUAAAAGAGAAACCCGAACUU ..(((((((.((....)).))))))).(((((..-------....((((.((..(((...----.))))).)))).......)).)))........................ ( -18.82) >DroYak_CAF1 11771 97 - 1 UGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAA-------AAAAACAUACGAAUACG--------UACGAAUGUGCACAAAGUCAACAAAUUAAAAGAGAAACCCAAACUU ..(((((((.((....)).))))))).(((((..-------....((((.((......--------..)).)))).......)).)))........................ ( -15.52) >consensus UGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAACAAAA__AAAAAAACACGAAUACGUACAUACGUACGAAUGUGUACAAAGCCAACAAAUUAAAAGAGAAACCCGAACUU ..(((((((.((....)).)))))))((((...................(((....)))......((((......))))............................)))). (-12.80 = -13.44 + 0.64)

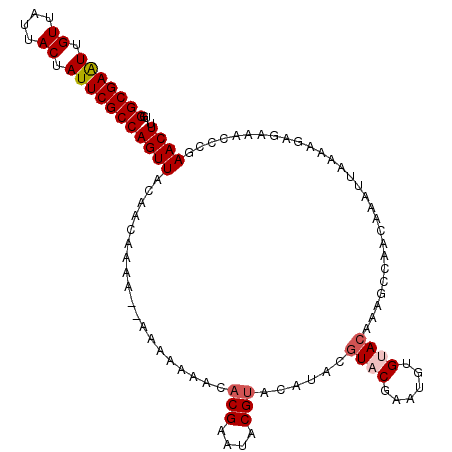
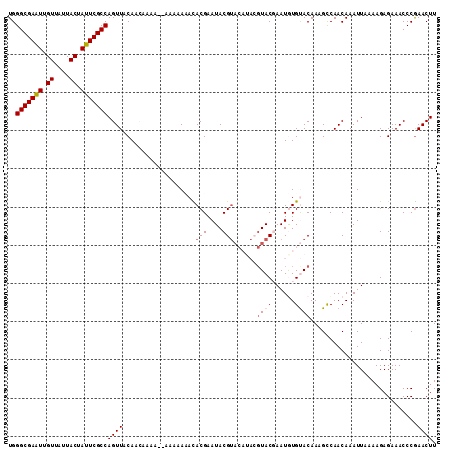
| Location | 1,802,965 – 1,803,060 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 67.74 |
| Mean single sequence MFE | -23.57 |
| Consensus MFE | -9.10 |
| Energy contribution | -10.60 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 1802965 95 - 22224390 -----AUUCGUGCCCGUGCCUGU----GUGUGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAACAAAAA---AAAAAUACACGAAUACGUACAUACGUACGAAUGU -----((((((((..((((.(((----.(((((((((((.((....)).))))))).((....)).....---.......)))).))).))))....)))))))).. ( -28.20) >DroSec_CAF1 11475 96 - 1 -----AUCCGCACCCGUGCCUGU----GUGUGGGCGAGUUGUUAUUACUAUUCGCCAGUUACAACAAAAAC--AAAAAAACACGAAUACGUACAUACGUACCAAUAU -----..........((((.(((----((((((((((((.((....)).))))))).(((........)))--...............)))))))).))))...... ( -22.10) >DroSim_CAF1 11555 92 - 1 -----AUCCGCGCCCGUGCCUGU----GUGUGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAACAAAA------AAAAACACGAAUACGUACAUACGUACCAAUAU -----....(((...((((.(((----.(((((((((((.((....)).))))))).((....))....------.....)))).))).))))...)))........ ( -21.90) >DroEre_CAF1 11307 94 - 1 CAUACAUAUGUAUCCGUGCCCGUGCCUGUGUGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAA---------AAAAACAUACGAAUACGUA----AGUACGAAUGU ..(((.(((((((.((((........((((..(((((((.((....)).)))))))...)))).---------.......)))).)))))))----.)))....... ( -25.39) >DroYak_CAF1 11808 86 - 1 CAUACAUUUGUAUCCGUGGCUGU----GUGUGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAA---------AAAAACAUACGAAUACG--------UACGAAUGU ...(((((((((..((((..(((----((((.(((((((.((....)).)))))))........---------....)))))))..))))--------))))))))) ( -28.92) >DroPer_CAF1 26459 93 - 1 -----AGUGUUGGCAUUGUAAUA----GUGUGUGCAAAUUGAGA-----AGGCCCCAAUUUCAUGAGCACCAAAAAAAGCAACGAAGAUCUGCAUACCCUAGAAAUG -----........((((....((----(.(((((((.(((..((-----((.......))))....((..........))......))).))))))).)))..)))) ( -14.90) >consensus _____AUUCGUACCCGUGCCUGU____GUGUGGGCGAAUUGUUAUUACUAUUCGCCAGUUACAACAAAA____AAAAAAACACGAAUACGUACAUACGUACGAAUGU ...............((((........(((..(((((((.((....)).)))))))...)))...................(((....)))......))))...... ( -9.10 = -10.60 + 1.50)
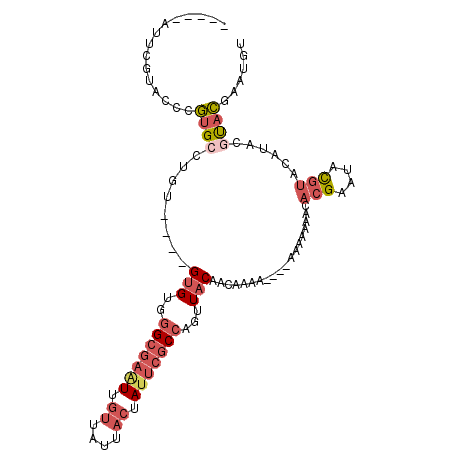
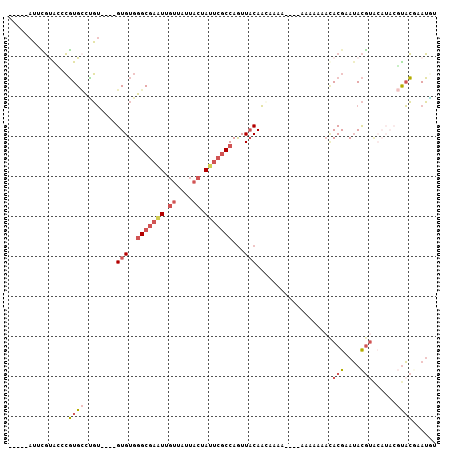
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:50:07 2006