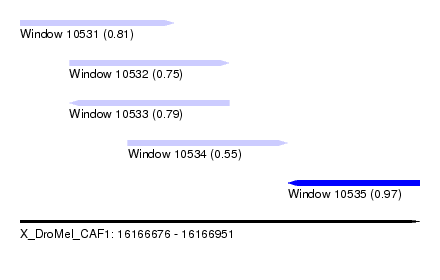
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,166,676 – 16,166,951 |
| Length | 275 |
| Max. P | 0.966252 |

| Location | 16,166,676 – 16,166,782 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 93.12 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -25.88 |
| Energy contribution | -27.12 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16166676 106 + 22224390 AUUCGAAUUGAAAGUACAGUCAGACGGGAA------CACACAAAAUGAAAAUGUGUAUGUGUAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUAUG ..((..((((......))))..))..((((------((((((((......((((((((((...(((((((...))))))))))))))))).))))))))))))......... ( -35.40) >DroSec_CAF1 6699 106 + 1 AUUCGAAUUGAAAGUACAGUCAGACGGGAA------CACACAAAAUGAAAAUGUGUAUGUGUAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUAUG ..((..((((......))))..))..((((------((((((((......((((((((((...(((((((...))))))))))))))))).))))))))))))......... ( -35.40) >DroSim_CAF1 7051 106 + 1 AUUCGAAUUGAAAGUACAGUCAGACGGGAA------CACACAAAAUGAAAAUGUGUAUGUGUAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUAUG ..((..((((......))))..))..((((------((((((((......((((((((((...(((((((...))))))))))))))))).))))))))))))......... ( -35.40) >DroEre_CAF1 7056 106 + 1 AUUCGAAUUGGAAGUACAGUCAGACGGGAACACAAACACACAAAAUGAAAAUGUG------UAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUUUA ..((..((((......))))..)).((((((((((((.((.....))...(((((------(((((.((((....)))).)))))))))))))))))).))))......... ( -29.90) >consensus AUUCGAAUUGAAAGUACAGUCAGACGGGAA______CACACAAAAUGAAAAUGUGUAUGUGUAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUAUG ..((..((((......))))..))..((((......((((((((......((((((((((...(((((((...))))))))))))))))).))))))))))))......... (-25.88 = -27.12 + 1.25)

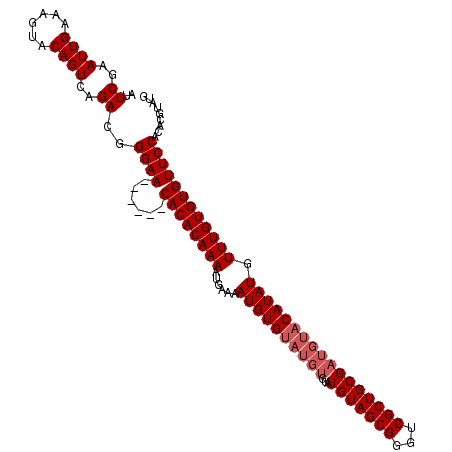

| Location | 16,166,710 – 16,166,820 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 93.51 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -27.65 |
| Energy contribution | -28.77 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16166710 110 + 22224390 CAAAAUGAAAAUGUGUAUGUGUAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACAUACAGCUGCGUUCGUGGAAAUAGCACUGA-AAGGC ......(((.((((((((((...(((((((...))))))))))))))))).)))((((.((((((((((((((....)))..)))))..))))))...))))...-..... ( -32.60) >DroSec_CAF1 6733 110 + 1 CAAAAUGAAAAUGUGUAUGUGUAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACUUACAGCUGCGUUCGUGGAAAUAGCACUGA-AAGGC ......(((.((((((((((...(((((((...))))))))))))))))).)))((((.((((((((((((((....)))..)))))..))))))...))))...-..... ( -32.60) >DroSim_CAF1 7085 110 + 1 CAAAAUGAAAAUGUGUAUGUGUAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACUUACAGCUGCGUUCGUGGAAAUAGCACUGA-AAGGC ......(((.((((((((((...(((((((...))))))))))))))))).)))((((.((((((((((((((....)))..)))))..))))))...))))...-..... ( -32.60) >DroYak_CAF1 7742 105 + 1 CAAAAUGAAAAUGUG------UAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACAUACAGCUGCGUUCGUGGGAAUAGUAGUGCGAGAGC ...........((((------(.(((((((...)))))))..((((((((((..(((.....))))))))))))))))))(((((((((....)))).)))))((....)) ( -33.10) >consensus CAAAAUGAAAAUGUGUAUGUGUAUGUAGCGGGUCGCUGCGAUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACAUACAGCUGCGUUCGUGGAAAUAGCACUGA_AAGGC ......(((.((((((((((...(((((((...))))))))))))))))).)))((((.((((((((((((((....)))..)))))..))))))...))))......... (-27.65 = -28.77 + 1.13)


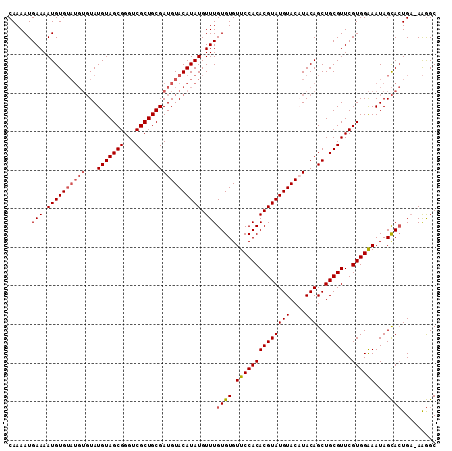
| Location | 16,166,710 – 16,166,820 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 93.51 |
| Mean single sequence MFE | -23.83 |
| Consensus MFE | -20.57 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16166710 110 - 22224390 GCCUU-UCAGUGCUAUUUCCACGAACGCAGCUGUAUGUACAUACGUGUGGAACACACAAACAUAUGUACAUCGCAGCGACCCGCUACAUACACAUACACAUUUUCAUUUUG ((..(-((.(((.......)))))).)).((((((((((((((..((((.....))))....))))))))).))))).................................. ( -25.60) >DroSec_CAF1 6733 110 - 1 GCCUU-UCAGUGCUAUUUCCACGAACGCAGCUGUAAGUACAUACGUGUGGAACACACAAACAUAUGUACAUCGCAGCGACCCGCUACAUACACAUACACAUUUUCAUUUUG ((..(-((.(((.......)))))).)).((((..((((((((..((((.....))))....))))))).)..)))).................................. ( -22.50) >DroSim_CAF1 7085 110 - 1 GCCUU-UCAGUGCUAUUUCCACGAACGCAGCUGUAAGUACAUACGUGUGGAACACACAAACAUAUGUACAUCGCAGCGACCCGCUACAUACACAUACACAUUUUCAUUUUG ((..(-((.(((.......)))))).)).((((..((((((((..((((.....))))....))))))).)..)))).................................. ( -22.50) >DroYak_CAF1 7742 105 - 1 GCUCUCGCACUACUAUUCCCACGAACGCAGCUGUAUGUACAUACGUGUGGAACACACAAACAUAUGUACAUCGCAGCGACCCGCUACAUA------CACAUUUUCAUUUUG ((..(((..............)))..)).((((((((((((((..((((.....))))....))))))))).))))).............------............... ( -24.74) >consensus GCCUU_UCAGUGCUAUUUCCACGAACGCAGCUGUAAGUACAUACGUGUGGAACACACAAACAUAUGUACAUCGCAGCGACCCGCUACAUACACAUACACAUUUUCAUUUUG ((........(((.............)))((((((((((((((..((((.....))))....))))))))).))))).....))........................... (-20.57 = -21.32 + 0.75)

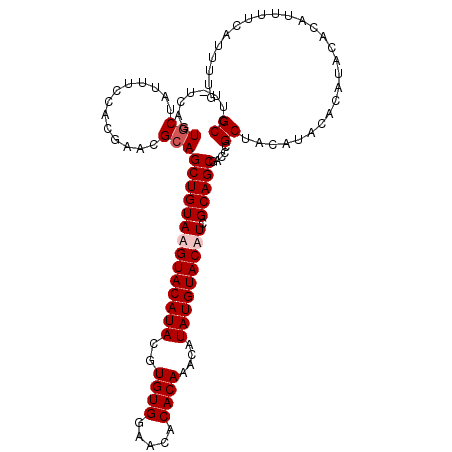

| Location | 16,166,750 – 16,166,860 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 89.29 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16166750 110 + 22224390 AUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACAUACAGCUGCGUUCGUGGAAAUAGCACUGA-AAGGCAUCAAACGGUUUUUUUCCGUAUUCCCUUUUGUUAAUUAAU ((((((((((((..(((.....)))))))))))))))...((((..((....))..))))...((-((((......((((.......))))....)))))).......... ( -27.70) >DroSec_CAF1 6773 108 + 1 AUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACUUACAGCUGCGUUCGUGGAAAUAGCACUGA-AAGGCAGCAAACGAUUUUAUUCUGAAUUCCCUUUGCUU--UGAAU ..((((((((((..(((.....))))))))))))).....(((((.((((((.......))).))-)..)))))..............................--..... ( -25.80) >DroSim_CAF1 7125 110 + 1 AUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACUUACAGCUGCGUUCGUGGAAAUAGCACUGA-AAGGCAGCAAACGAUUUUAUUCUGAAUUCCAUUUUGUUAAUGAAU ..((((((((((..(((.....))))))))))))).....(((((.((((((.......))).))-)..))))).....................((((.....))))... ( -26.50) >DroYak_CAF1 7776 109 + 1 AUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACAUACAGCUGCGUUCGUGGGAAUAGUAGUGCGAGAGCAUCAAACGGUU-U-UUUCGUAUUCCCUUUUGUUAAUUAAU ((((((((((((..(((.....)))))))))))))))..............((((((....((((....))))...((((..-.-..)))))))))).............. ( -30.50) >consensus AUGUACAUAUGUUUGUGUGUUCCACACGUAUGUACAUACAGCUGCGUUCGUGGAAAUAGCACUGA_AAGGCAGCAAACGAUUUUAUUCCGAAUUCCCUUUUGUUAAUGAAU ..((((((((((..(((.....))))))))))))).....(((((.((.(((.......)))....)).)))))..................................... (-19.12 = -19.50 + 0.38)


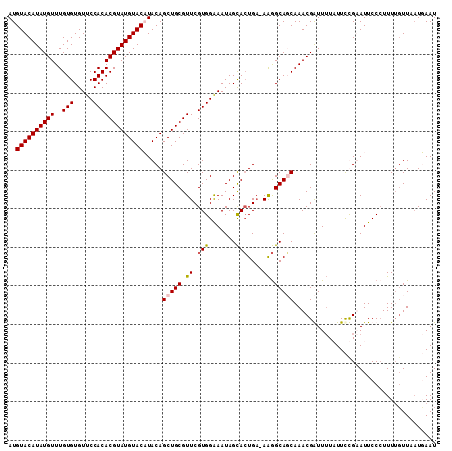
| Location | 16,166,860 – 16,166,951 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 90.30 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -20.85 |
| Energy contribution | -20.97 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16166860 91 - 22224390 AUAUGUUUUGUUAAAGAGCAGUGCAGGUACAAAAAGCCAUUUGUAACUGCUUUGCUUGUUUUGCGACACAGAAGACGAAGCGAAAAAAAAA ...(((((((((...((((((.((((.((((((......)))))).)))).)))))).(((((.....))))))))))))))......... ( -26.70) >DroSec_CAF1 6881 86 - 1 AUAUGUUUUGUUAAAAAACAGUGCAGGUACUAAAAGCCAUUUGUAACUGCAUUGCUUGUUUUGGGACACAGAAGACGAAGCAA-----AGU ...(((((((((......((((((((.(((............))).))))))))....(((((.....)))))))))))))).-----... ( -23.40) >DroSim_CAF1 7235 86 - 1 AUAUGUUUUGUUAAAAAACAGUGCAGGUACUAAAAGCCAUUUGUAACUGCAUUGCUUGUUUUGCGACACAGAAGACGAAGCAA-----AGU ...(((((((((......((((((((.(((............))).))))))))....(((((.....)))))))))))))).-----... ( -22.10) >consensus AUAUGUUUUGUUAAAAAACAGUGCAGGUACUAAAAGCCAUUUGUAACUGCAUUGCUUGUUUUGCGACACAGAAGACGAAGCAA_____AGU ...(((((((((......((((((((.(((............))).))))))))....(((((.....))))))))))))))......... (-20.85 = -20.97 + 0.11)

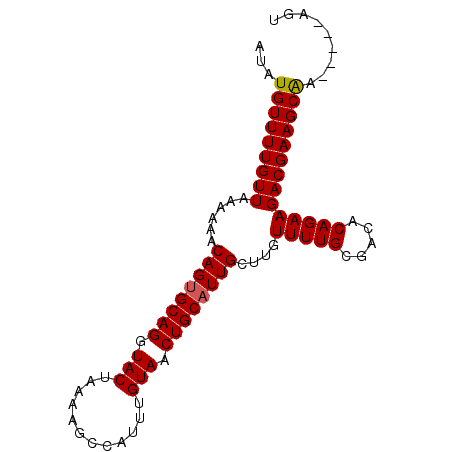
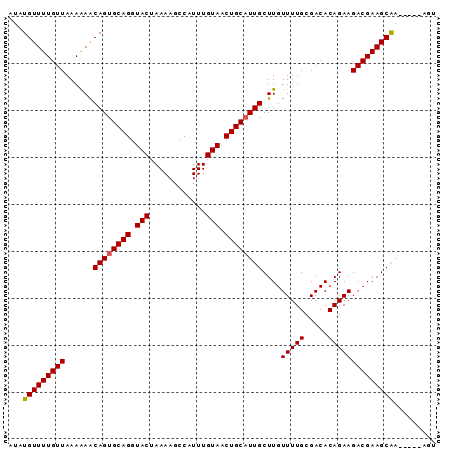
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:14:06 2006