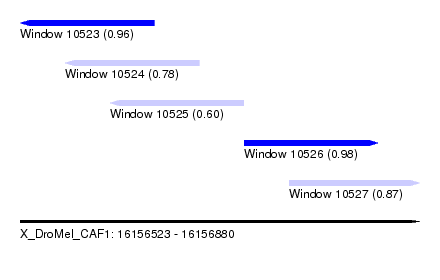
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,156,523 – 16,156,880 |
| Length | 357 |
| Max. P | 0.980143 |

| Location | 16,156,523 – 16,156,643 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -43.66 |
| Consensus MFE | -42.94 |
| Energy contribution | -42.34 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16156523 120 - 22224390 AGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCAGGAGAUACAGGAAGCGUCGCCCAAGGGCGAACUGGAUGUGGUCUUCAACAAGUACUGCAGCCGGCGUUCGAAUGCCGUCG (((((.......((((((((........))))))))....(((((((((...((..(((((....))))).))...))).)))))).....)))))...(.((((((....)))))).). ( -43.10) >DroSec_CAF1 2465 120 - 1 AGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCAGGAAAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUGGUUUUCAACAAGUACUGCAGCCGGCGUUCCAAUGCUGUCG ...(((.....)))((((((........))))))(((((((......).((((.(((((((....))))).((((.((..((...........))..)).))))))))))..)))))).. ( -40.50) >DroSim_CAF1 3241 120 - 1 AGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCAGGAAAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUGGUUUUCAACAAGUACUGCAGCCGGCGUUCCAAUGCCGUCG ((((((((....((((((((........))))))))....(......)..))))).(((((....))))))))((.((..((...........))..)).))(((((....))))).... ( -42.80) >DroEre_CAF1 2410 120 - 1 AGUGCCUCAAUGGCAUUGUCAACUACACGGCGAUGCAGCAGGAGAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUGGUCUUCAACAAGUACUGCAGCCGGCGUUCGAAUGCCGUGG ((((((.....))))))........(((((((((((..(..(.....)..)..)))))(((....)))(((((((.((..(((((....))).))..)).)))..))))....)))))). ( -45.40) >DroYak_CAF1 3289 120 - 1 AGUGCCUCAAUGGCAUCGUCAACUACACGGCGAUGCAGCAGGAGAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUGGUCUUUAACAAGUACUGCAGCCGGCGUUCGAAUGCCGUGG ((((((((....((((((((........))))))))....(......)..))))).(((((....))))))))((.((..(((((....))).))..)).))(((((....))))).... ( -46.50) >consensus AGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCAGGAGAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUGGUCUUCAACAAGUACUGCAGCCGGCGUUCGAAUGCCGUCG ((((((((....((((((((........))))))))....(......)..))))).(((((....))))))))((.((..(((((....))).))..)).))(((((....))))).... (-42.94 = -42.34 + -0.60)



| Location | 16,156,563 – 16,156,683 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -44.04 |
| Consensus MFE | -39.20 |
| Energy contribution | -38.64 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16156563 120 - 22224390 GCAUACGGCGAGAUCGAGUCCGGAUUCAUGGUGCUAACCGAGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCAGGAGAUACAGGAAGCGUCGCCCAAGGGCGAACUGGAUGUG ((.....))........((((((.(((..(..(((..((..(((.(((....((((((((........)))))))).....))).))).)).)))..)(((....))))))))))))... ( -41.20) >DroSec_CAF1 2505 120 - 1 GCGUACGGUGAGAUCGAGUCCGGAUUCAUGGUGCUAACCGAGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCAGGAAAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUG ..((((.(((((.(((....))).))))).))))...((.((((((((....((((((((........))))))))....(......)..))))).(((((....))))))))))..... ( -45.60) >DroSim_CAF1 3281 120 - 1 GCGUACGGUGAGAUCGAGUCCGGAUUCAUGGUGCUAACCGAGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCAGGAAAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUG ..((((.(((((.(((....))).))))).))))...((.((((((((....((((((((........))))))))....(......)..))))).(((((....))))))))))..... ( -45.60) >DroEre_CAF1 2450 120 - 1 GCGUACGGCGAGAUCGAGUCCGGAUUCAUGGUGCUGACGGAGUGCCUCAAUGGCAUUGUCAACUACACGGCGAUGCAGCAGGAGAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUG .(((.(((((..((.((((....))))))..)))))))).((((((((....((((((((........))))))))....(......)..))))).(((((....))))))))....... ( -41.40) >DroYak_CAF1 3329 120 - 1 GCGUUCGGCGAGAUCGAAUCCGGAUUUAUGGUGCUAACCGAGUGCCUCAAUGGCAUCGUCAACUACACGGCGAUGCAGCAGGAGAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUG ((((((((......((..(((.......((((....)))).(((.(((....((((((((........)))))))).....))).))).)))..))(((((....))))).)))))))). ( -46.40) >consensus GCGUACGGCGAGAUCGAGUCCGGAUUCAUGGUGCUAACCGAGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCAGGAGAUACAGGAGGCGUCGCCCAAGGGCGAACUGGAUGUG ((.....))........((((((((((..(((....)))))))(((((....((((((((........))))))))....(......)..))))).(((((....))))).))))))... (-39.20 = -38.64 + -0.56)


| Location | 16,156,603 – 16,156,723 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -47.12 |
| Consensus MFE | -43.62 |
| Energy contribution | -43.14 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16156603 120 - 22224390 CAAAUGCAUCGAGGUGGCCGGCGAGGAGGCGGGCGUCGAGGCAUACGGCGAGAUCGAGUCCGGAUUCAUGGUGCUAACCGAGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCA ....(((.....(..(.(((.(.....).))).)..)(((((((.(((..(((((((((....))))..))).))..))).)))))))....((((((((........)))))))).))) ( -45.50) >DroSec_CAF1 2545 120 - 1 CAAAUGCAUCGAGGUGGCCGGCGAGGAGGCGGGCGGCGAGGCGUACGGUGAGAUCGAGUCCGGAUUCAUGGUGCUAACCGAGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCA ....(((......((.(((.((......)).))).))((((((..((((.(((((((((....))))..))).)).))))..))))))....((((((((........)))))))).))) ( -47.20) >DroSim_CAF1 3321 120 - 1 CAAAUGCAUCGAGGUGGCCGGCGAGGAGGCGGGCGGCGAGGCGUACGGUGAGAUCGAGUCCGGAUUCAUGGUGCUAACCGAGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCA ....(((......((.(((.((......)).))).))((((((..((((.(((((((((....))))..))).)).))))..))))))....((((((((........)))))))).))) ( -47.20) >DroEre_CAF1 2490 120 - 1 CAAAUGCGUCGAGGUGGCCGGCGAGGAGGCGGGCGGCGAGGCGUACGGCGAGAUCGAGUCCGGAUUCAUGGUGCUGACGGAGUGCCUCAAUGGCAUUGUCAACUACACGGCGAUGCAGCA ....(((((((..((((((.((......)).)))(((....(((.(((((..((.((((....))))))..)))))))).((((((.....))))))))).....)))..)))))))... ( -47.20) >DroYak_CAF1 3369 120 - 1 CAAAUGCAUCGAGGUGGCCGGCGAGGAGGCGGGCGGCGAGGCGUUCGGCGAGAUCGAAUCCGGAUUUAUGGUGCUAACCGAGUGCCUCAAUGGCAUCGUCAACUACACGGCGAUGCAGCA ....(((......((.(((.((......)).))).))(((((((((((..(((((((((....))))..))).))..)))))))))))....((((((((........)))))))).))) ( -48.50) >consensus CAAAUGCAUCGAGGUGGCCGGCGAGGAGGCGGGCGGCGAGGCGUACGGCGAGAUCGAGUCCGGAUUCAUGGUGCUAACCGAGUGCCUCAAUGGCAUUGUCAACUACACGGCAAUGCAGCA ....(((......((.(((.((......)).))).))((((((..(((..(((((((((....))))..))).))..)))..))))))....((((((((........)))))))).))) (-43.62 = -43.14 + -0.48)

| Location | 16,156,723 – 16,156,843 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -48.76 |
| Consensus MFE | -40.00 |
| Energy contribution | -40.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16156723 120 + 22224390 UUGCGAAAUAGCUCCUUGGCGUCGUUCACGCUAAAGUUUGACUUCCUCAGCUGGUCGGGCAGCUGGGUCUGGAUCUGAUCCAGGUCUAGCUGGGCCAUUAUGCCGGCAGUGGCGAAUAUA (((((((..((((..(((((((.....)))))))))))....)))....((((((..(((((((((..((((((...))))))..))))))..))).....))))))....))))..... ( -46.40) >DroSec_CAF1 2665 120 + 1 UUGCGAAAUAGCUCCUUGGCGUCAUUCACGCUAAAGUUCGAGUUGCUUAGCUGGUCGGGCAGCUGGGUCUGGAUCUGAUCCAGGUCUAGCUGGGCCAUUACGCCGGCAGUGGCGAGUAUA ..........((((.(((((((.....))))))).......((..((..((((((..(((((((((..((((((...))))))..))))))..))).....))))))))..))))))... ( -50.40) >DroSim_CAF1 3441 120 + 1 UUGCGAAAUAGCUCCUUGGCGUCAUUCACGCUAAAGUUCGACUUGGUUAGCUGGUCGGGCAGCUGAGUCUGGAUCUGCUCAAGGUCUAGCUGGGCCAUUACGCCGGCAGUGGAGAGUAUA ...........(((((((((((.....)))))))...............(((((.((....(((.((.((((((((.....)))))))))).))).....)))))))...))))...... ( -40.90) >DroEre_CAF1 2610 120 + 1 UUGCGGAUUAGCUCCUUGGCGUCGUUGACGCUAAAGUUCGACUUGCUCAGCUGGUCGGGCAGUUGGGUCUGGAUCUGAUCCAGGUCCAGCUGGGCCACUAUGCCAGGAGUUGCCAGUACA .(((((..(((((((((((((((...)))))))).((((((((.((...)).))))))))((((((..((((((...))))))..)))))).(((......))).))))))))).))).. ( -55.20) >DroYak_CAF1 3489 120 + 1 UUGCGGAUUAGCUCCUUGGCGUCGUUAACGCUAAAGUUUGACUUGCUCAGCUGGUCGGGCAGUUGGGUCUGGAUCUGGUCCAGGUCCAGCUGGGCCAUUAUGCCAGCCGCUGCCAGUACC ..(((..((((((..(((((((.....))))))))).))))..)))...(((((((((.(((((((..((((((...))))))..)))))))(((......)))..)))..))))))... ( -50.90) >consensus UUGCGAAAUAGCUCCUUGGCGUCGUUCACGCUAAAGUUCGACUUGCUCAGCUGGUCGGGCAGCUGGGUCUGGAUCUGAUCCAGGUCUAGCUGGGCCAUUAUGCCGGCAGUGGCGAGUAUA ..(((.(((.((((.(((((((.....))))))).((((((((.(.....).))))))))((((((..(((((.....)))))..)))))))))).))).)))................. (-40.00 = -40.24 + 0.24)



| Location | 16,156,763 – 16,156,880 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -44.70 |
| Consensus MFE | -35.66 |
| Energy contribution | -37.94 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16156763 117 + 22224390 ACUUCCUCAGCUGGUCGGGCAGCUGGGUCUGGAUCUGAUCCAGGUCUAGCUGGGCCAUUAUGCCGGCAGUGGCGAAUAUAACCAGCCAAAUGGCGUAUAUCGCAUGGCUUACAUA---UG .........((((((..(((((((((..((((((...))))))..))))))..))).....))))))(((.((((.((((....(((....))).))))))))...)))......---.. ( -46.80) >DroSec_CAF1 2705 117 + 1 AGUUGCUUAGCUGGUCGGGCAGCUGGGUCUGGAUCUGAUCCAGGUCUAGCUGGGCCAUUACGCCGGCAGUGGCGAGUAUAACCAACCAAAUGGCGCAUAUCGCAUGGCUUACAUA---UA .((..((..((((((..(((((((((..((((((...))))))..))))))..))).....))))))))..))..(((...(((.((....)).((.....)).)))..)))...---.. ( -45.70) >DroSim_CAF1 3481 117 + 1 ACUUGGUUAGCUGGUCGGGCAGCUGAGUCUGGAUCUGCUCAAGGUCUAGCUGGGCCAUUACGCCGGCAGUGGAGAGUAUAACCAACCAAAUGGCGCAUAUCGCAUGGCUUACAUA---UA ..((((((.(((((.((....(((.((.((((((((.....)))))))))).))).....)))))))...((.........))))))))...(((.....)))(((.....))).---.. ( -36.60) >DroEre_CAF1 2650 120 + 1 ACUUGCUCAGCUGGUCGGGCAGUUGGGUCUGGAUCUGAUCCAGGUCCAGCUGGGCCACUAUGCCAGGAGUUGCCAGUACAACCAGCCAAAUGGCACAUAUCGCAUGGCGUACAUACAGUG .........((((...((.(((((((..((((((...))))))..)))))).).))..((((((((((((((......))))..(((....))).....)).).)))))))....)))). ( -45.50) >DroYak_CAF1 3529 117 + 1 ACUUGCUCAGCUGGUCGGGCAGUUGGGUCUGGAUCUGGUCCAGGUCCAGCUGGGCCAUUAUGCCAGCCGCUGCCAGUACCACCAGCCAAAUGGCGCAUAUCGCUUGGCUUACAUA---UG .........(((((((((.(((((((..((((((...))))))..)))))))(((......)))..)))..))))))......(((((...((((.....)))))))))......---.. ( -48.90) >consensus ACUUGCUCAGCUGGUCGGGCAGCUGGGUCUGGAUCUGAUCCAGGUCUAGCUGGGCCAUUAUGCCGGCAGUGGCGAGUAUAACCAGCCAAAUGGCGCAUAUCGCAUGGCUUACAUA___UG (((((((..((((((..(((((((((..(((((.....)))))..))))))..))).....))))))...))))))).......((((....(((.....))).))))............ (-35.66 = -37.94 + 2.28)

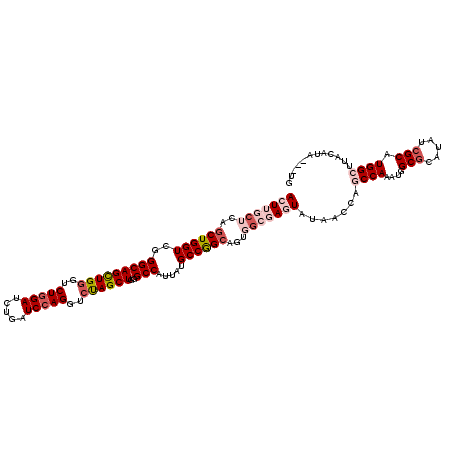
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:59 2006