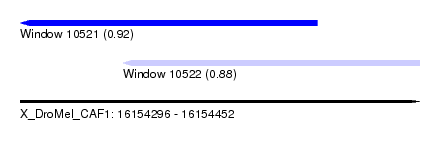
| Sequence ID | X_DroMel_CAF1 |
|---|---|
| Location | 16,154,296 – 16,154,452 |
| Length | 156 |
| Max. P | 0.919958 |

| Location | 16,154,296 – 16,154,412 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.41 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -31.76 |
| Energy contribution | -31.80 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16154296 116 - 22224390 AAUG-GGCAGCACAAAGUGCGUUUAUGGGCGCCCUAAAGUGAUGUAACAAUUGCUGCGACCGCAGCA---GCAGCGAAACGCGCGGAAUGGAACGCACGGAACGAUAUCGUAGUAUCGUA ...(-(((.((((...))))(((....)))))))....(((.(((..((..((((((....))))))---((.(((...)))))....))..))))))...(((((((....))))))). ( -38.90) >DroSec_CAF1 150 117 - 1 AAUGGGGCAGCAAAAAGUGCGUUUAUGGGCGCCCUAAAGUGAUGUAACAAUUGCUGCGACCGCAGCA---GCGGCGAAACGCGCGGAAUGGAACGCACGGAACGAUAUCGUAGUAUCGUA ..((((((.(((.....)))(((....)))))))))..(((.(((..((..((((((....))))))---(((......)))......))..))))))...(((((((....))))))). ( -40.30) >DroSim_CAF1 935 116 - 1 AAUG-GGCAGCAUAAAGUGCGUUUAUGGGCGCCCUAAAGUGAUGUAACAAUUGCUGCGACCGCAGCA---GCGGCGAAACGCGCGGAAUGGAACGCACGGAACGAUAUCGUAGUAUCGUA ...(-(((.((((...))))(((....)))))))....(((.(((..((..((((((....))))))---(((......)))......))..))))))...(((((((....))))))). ( -38.30) >DroEre_CAF1 149 113 - 1 AAUG-GGCCGCGAAAAGUGCGUUCAUGGGCGCCGCAAAGUGAUGUAACAAUAGCUGCGACCGCAG------CAGCGGAACGCGCGGAAUGGAACGCACGGAACGGUAUCGUAGUAUCGUA .(((-(((((......((((((((.((.(((((((...((......))....(((((....))))------).))))..))).)).....))))))))....)))).))))......... ( -42.30) >DroYak_CAF1 1032 119 - 1 AAUG-GACCGCAAAAAGUGCGUUUAUGGGCGCCGCAAAGUGAUGUAACAAUAGCUGCGACCGCAGCAGCAGCAGCGGAACGGGCGGAAUAGAACGCAUGGAACGGUAUCGUAGUAUCGUA ..((-..((((.....(.(((((....))))))((...((......))....(((((....))))).))....))))..)).(((........))).....(((((((....))))))). ( -35.80) >consensus AAUG_GGCAGCAAAAAGUGCGUUUAUGGGCGCCCUAAAGUGAUGUAACAAUUGCUGCGACCGCAGCA___GCAGCGAAACGCGCGGAAUGGAACGCACGGAACGAUAUCGUAGUAUCGUA ................((((((((.....(((......((..(((......((((((....))))))......)))..))..))).....))))))))...(((((((....))))))). (-31.76 = -31.80 + 0.04)



| Location | 16,154,336 – 16,154,452 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -46.44 |
| Consensus MFE | -31.72 |
| Energy contribution | -33.52 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>X_DroMel_CAF1 16154336 116 - 22224390 AGGGCGCAUGAACGCGACCAUUUAUCCGUUUUGAUGGGGGAAUG-GGCAGCACAAAGUGCGUUUAUGGGCGCCCUAAAGUGAUGUAACAAUUGCUGCGACCGCAGCA---GCAGCGAAAC (((((((((((((((..((((((..((((....))))..)))))-))).((((...))))))))))..)))))))...((......))..(((((((....))))))---)......... ( -45.30) >DroSec_CAF1 190 117 - 1 AGGGUGCAUGAACGCGACCAUUUAACCGUUUUGGUGGGGGAAUGGGGCAGCAAAAAGUGCGUUUAUGGGCGCCCUAAAGUGAUGUAACAAUUGCUGCGACCGCAGCA---GCGGCGAAAC (((((((((((((((..((((((..(((......)))..)))))).)).(((.....)))))))))..)))))))..((..((......))..)).((.((((....---)))))).... ( -44.70) >DroSim_CAF1 975 116 - 1 AGGCCGCAUGAACGCGACCAUUUAACCGUUUUGGUGGGGGAAUG-GGCAGCAUAAAGUGCGUUUAUGGGCGCCCUAAAGUGAUGUAACAAUUGCUGCGACCGCAGCA---GCGGCGAAAC ..(((.(((((((((..((((((..(((......)))..)))))-)((........))))))))))))))(((.....((......))..(((((((....))))))---).)))..... ( -45.20) >DroEre_CAF1 189 113 - 1 AAGGCGCAUGAACGCGCCCAUUUAUCCGUUUUGGCCGGGGAAUG-GGCCGCGAAAAGUGCGUUCAUGGGCGCCGCAAAGUGAUGUAACAAUAGCUGCGACCGCAG------CAGCGGAAC ...((.(((((((((((((((((.((((.......)))))))))-))).((.....))))))))))).)).((((...((......))....(((((....))))------).))))... ( -52.70) >DroYak_CAF1 1072 119 - 1 UAGGCGCAUGAACGCGACCAUUUAUGCGUGCUGGGCGGGGAAUG-GACCGCAAAAAGUGCGUUUAUGGGCGCCGCAAAGUGAUGUAACAAUAGCUGCGACCGCAGCAGCAGCAGCGGAAC ...((.((((((((((........)))(..((..((((......-..))))....))..)))))))).)).((((...((......))....(((((....))))).......))))... ( -44.30) >consensus AGGGCGCAUGAACGCGACCAUUUAUCCGUUUUGGUGGGGGAAUG_GGCAGCAAAAAGUGCGUUUAUGGGCGCCCUAAAGUGAUGUAACAAUUGCUGCGACCGCAGCA___GCAGCGAAAC (((((((((((((((..((((((..((((....))))..))))).)...((.....))))))))))..)))))))...((..(((......((((((....))))))......)))..)) (-31.72 = -33.52 + 1.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:13:55 2006